Circulating Cell-Free DNA and Systemic Inflammatory Response after Self-Expandable Metal Stent for Malignant Bowel Obstruction
Malene Broholm*, 1, 2, Rasmus Bojesen1, 3, Mikail Gögenur1, Andreas Weinberger Rosen1, Sara Watt1, Mustafa Bulut1, 4, Rasmus Vogelsang1, Henning Quist Jensen3, Adile Orhan1, Ellen Bjerrum1, Christina Søs Auður Andersen5, Niels Pallisgaard5, Thomas Litman6, Jesper T. Troelsen2, Ismail Gögenur1, 4
1Center for Surgical Science, Department of Surgery, Zealand University Hospital, Denmark
2Department of Science and Environment, Roskilde University, Denmark
3Department of Surgery, Slagelse Hospital, Denmark
4Department of Clinical Medicine, University of Copenhagen, Denmark
5Department of Pathology, Zealand University Hospital, Denmark
6Department of International Health, Immunology and Microbiology, University of Copenhagen, Denmark
*Corresponding author: Malene Broholm, Center for Surgical Science, Department of Surgery, Zealand University Hospital, Denmark.
Received: 28 September 2022; Accepted: 06 October 2022; Published: 16 November 2022
Article Information
Citation: Malene Broholm, Rasmus Bojesen, Mikail Gögenur, Andreas Weinberger Rosen, Sara Watt, Mustafa Bulut, Rasmus Vogelsang, Henning Quist Jensen, Adile Orhan, Ellen Bjerrum, Christina Søs Auður Andersen, Niels Pallisgaard, Thomas Litman, Jesper T Troelsen, Ismail Gögenur. Circulating Cell-Free DNA and Systemic Inflammatory Response after Self-Expandable Metal Stent for Malignant Bowel Obstruction. Archives of Microbiology and Immunology 6 (2022): 247-255.
View / Download Pdf Share at FacebookAbstract
Purpose: A colonic stent as a bridge to elective surgery can improve short-term clinical outcomes. However, the placement of a stent may induce adverse effects on the immune system through the upregulation of proteins associated with migration, angiogenesis, and inflammation, which may affect the long-term oncological outcome. This study aimed to investigate systemic alterations in the systemic inflammatory response and circulating cell-free DNA in relation to stent placement.
Methods: This prospective observational study included 20 patients with acute malignant colonic obstruction. Blood samples were collected at baseline and 1, 4, and 24 hours after stent placement. Protein quantification was performed using a standardized proteomics panel, and baseline samples were compared with samples collected 24 hours after stent placement. Quantitative analysis of circulating cell-free DNA was evaluated at baseline and 1, 4, and 24 hours after stent placement.
Results: We identified 11 significantly changed protein concentrations (ARG1, GZMA, PGF, GZMH, KLRD1, CCL23, IL5, CCL19, PTN, MUC-16, CD8A) and a tendency towards higher concentration in three proteins (CAIX, CXCL1, IL6) in the 24h samples compared to baseline. Among the proteins mentioned, eight have been associated with tumor-promoting functions, such as proliferation, migration, and angiogenesis. Additionally, two proteins (CD8A and CCL19) are associated with anti-tumor effects. Quantitative analysis of circulating cell-free DNA did not reveal any significant differences between baseline, 1, 4, and 24 hours after stent placement.
Conclusions: Stent placement may induce a systemic inflammatory response potentially associated with tumor-promoting functions. However, we did not find an increase in systemic cell-free DNA. This study supports the importance of further investigating to determine systemic effects in relati
Keywords
<p>Colonic stent, SEMS, inflammatory response, cell free DNA</p>
Article Details
1. Introduction
Colorectal cancer (CRC) is one of the most commonly diagnosed cancers and one of the leading causes of cancer-related deaths, particularly in the economically developed world [1]. In 2020, approximately 1.9 million new colon and rectal cancer cases were diagnosed. In addition, it has been reported that 8-29% with primary CRC present with an acute malignant obstruction [2]. Acute malignant colonic obstruction is a life-threatening condition that requires immediate attention to improve the patient’s clinical condition [3-6]. Self-expanding metallic stent (SEMS) is used as an alternative to emergency surgery for acute malignant obstruction. SEMS can restore the luminal patency and thereby provides the opportunity to improve the clinical condition before an elective surgical resection (bridge to surgery). For short-term outcomes, studies have shown improved clinical outcomes such as lower morbidity rates, lower permanent stoma rates, and higher rates of primary anastomosis after SEMS as a bridge to surgery compared with those after emergency surgery [7, 8].
The European Society of Gastrointestinal Endoscopy (ESGE) guidelines published in 2020 strongly recommend SEMS as a palliative treatment to improve short-term clinical outcomes. However, the oncological outcomes after SEMS remain unclear, and ESGE advises SEMS as a bridge to surgery to be discussed in the individual cases [9]. A recent meta-analysis revealed higher overall and systemic recurrence rates after SEMS as a bridge to surgery compared with emergency surgery [7]. Mechanical compression may cause tissue damage, promote a more invasive cancer phenotype [10], and induce angiogenesis [11]. A recent study evaluating the systemic biomarkers in patients with SEMS placement found elevated levels of circulating cell-free DNA (cfDNA) and circulating tumor DNA (ctDNA) on day seven after SEMS placement [12], and high levels have been correlated with impaired prognosis in CRC [13, 14].
It is well known that cancer surgery induces a systemic stress response, and the inflammatory response associated with surgical trauma may increase the risk of recurrence in CRC [15-17]. SEMS placement may cause tissue damage. The additional persistent mechanical pressure from the SEMS may also induce an inflammatory response, which may affect the long-term oncological outcome of patients. We aimed to study indicators of the systemic inflammatory response through analyses of circulating cfDNA and a proteomic panel in blood prospectively before and after SEMS placement in patients with obstructing CRC.
2. Materials and Methods
This was a prospective observational study investigating the early systemic effects of SEMS. Patients were included between January 2018 and November 2019, from the Departments of Surgery, Zealand University Hospital, and Slagelse Hospital, Denmark. We planned to include 20 patients with CRC admitted to the surgical department due to malignant obstruction and treated with SEMS placement as either a palliative procedure or as a bridge to intended curative surgery. Patients with a previous SEMS insertion treated with a stent-in-stent were not eligible for inclusion. The study was explorative as no previous evidence on the systemic response after SEMS has been reported and no power calculation was made. The study was approved by the Ethics Committee in Region Zealand, Denmark (SJ-512) and the Danish Data Protection Agency (REG-059-2017). All patients signed informed consent forms before participating in the study.
2.1 Stenting Technique
SEMS insertions were performed by a combined endoscopic and fluoroscopic approach according to a standard technique as previously described [18, 19]. All procedures were performed by specialized endoscopists and were carried out under general anesthesia or sedation. A guidewire was introduced through the stenosis, and a stent was deployed over the guide wire. Finally, the correct position of the stent was confirmed by fluoroscopy. The colonic stents used were Wallflex (Boston Scientific, MA, USA), Evolution (Cook Medical, IN, USA), and Hanarostent (M.I. Tech., Seoul, Korea).
2.2 Blood samples
Baseline blood samples were collected on the day of SEMS placement, and follow-up samples were collected at 1, 4, and 24 hours after SEMS placement. All samples were collected and handled according to a standardized protocol. Samples were collected in 2 x 10 ml EDTA vacutainers. After collecting the samples, they were centrifuged at 3000g for 10 mins within one and a half hours after collection. Plasma was then transferred to a 15 ml tube (BD Falcon) and centrifuged for 10 min. Plasma was then divided into 2 x 5ml and 2 x 2ml vials and stored at -800C until analysis.
2.3 Inflammatory Response
Early systemic response to SEMS placement was characterized through the evaluation of protein biomarkers using a validated proteomics panel. The Immune-Oncology panel is a multiplex immunoassay targeting 92 protein biomarkers associated with promoting and inhibiting tumor immunity, chemotaxis, vascular and tissue remodeling, apoptosis, metabolism, and autophagy [20].
2.3.1 Proximity Extension Assay
Relative protein abundance was quantified using Proximity Extension Assay (PEA) from Olink Proteomics AB, Sweden [21]. The analysis was performed at BioXpedia A/S, Aarhus, Denmark. In brief, 1 µL of a sample was incubated with 92 antibody pairs labeled with DNA oligonucleotides. The pair of oligonucleotide-labeled antibodies bind to each of the 92 target proteins, after which dual binding in the proximity of matching antibody-probes results in hybridization of oligos, and subsequently, a PCR target sequence was formed. Finally, the target sequence was detected and quantified using standard real-time PCR. The 40 plasma samples were distributed randomly on the Fluidigm chip.
The PEA readout is Normalized Protein Expression (NPX), which is an arbitrary unit on a log2 scale, where a high NPX corresponds to a high protein abundance. External and internal controls are included in the PEA assay to monitor assay performance and adjust for intra- and inter-run variation. Four internal controls are added to each sample to monitor the quality of assay performance in each step, i.e. antibody binding, extension, and detection. Negative controls and inter-plate positive controls are run in triplicate on each plate [21]. Assay specific limit of detection was calculated as 3 times the standard deviation over the background signal. Normalization between plates was performed using intensity normalization.
2.3.2 Quantification of circulating cell-free DNA (cfDNA)
Extraction of plasma cfDNA was performed using a Perkin Elmer Chemagic 360 Robot, Waltham, Massachusetts, USA, with a CMG-1304 kit according to the manufacturer’s recommendations. All samples were analyzed using digital droplet PCR (ddPCR) QX-200 system from Bio-Rad, Hercules, California, USA. Levels of spike-in control were measured together with levels of immunoglobulin gene rearrangement as a control for potential contaminating lymphocyte DNA [22]. Further, a fragmentation ratio analysis was performed by measuring levels of 65 base pairs (bp) and 250 bp fragments of the EMC7 housekeeping gene. The EMC7 65 bp assay was also used to quantify the total levels of cfDNA.
2.4 Statistical analysis
A Shapiro-Wilk test was used to test the cfDNA concentration at the different time points for normality. A Friedman’s test was used to see if cfDNA concentration differed significantly between time points. Patients with missing data at any time point were excluded from the analysis. Friedman’s test is an alternative to the repeated measures ANOVA, when the assumption ofnormalityor equality of variance is not met. Differentially expressed proteins were identified by a paired t-test (patient eliminated as a factor, p<0.10), and significance were corrected for multiple testing by estimation of false discovery rate [23]. The data were visualized in Qlucore Omics Explorer v.3.7 (Qlucore AB, Lund, Sweden) by principal component analysis, heat maps, and unsupervised hierarchical clustering. All analyses were performed using R v4.1.
3. Results
Between January 2018 and December 2019, a total of 20 patients with obstructing CRC were enrolled in the study at Zealand University Hospital and Slagelse Hospital, Denmark. Among the included patients, nine underwent SEMS placement as a definitive palliative treatment, and eleven patients underwent SEMS placement as a bridge to elective surgery. The patient characteristics are presented in Table 1.
Table 1: Characteristics of patients undergoing colorectal stent for malignant obstruction
|
Characteristics |
N = 20 (%) |
|
Sex, male |
10 (50) |
|
Age, median (range) |
73 (57–88) |
|
Tumor location |
|
|
Ascending colon |
4 (20) |
|
Transverse colon |
2 (10) |
|
Descending colon |
3 (15 |
|
Sigmoid colon |
10 (50) |
|
Rectum |
1 (5) |
|
Tumor stage |
|
|
1 |
0 (0) |
|
2 |
4 (20) |
|
3 |
14 (70) |
|
4 |
2 (10) |
|
ASA score |
|
|
1 |
1 (5) |
|
2 |
15 (75) |
|
3 |
4 (20) |
|
Performance status |
|
|
0 |
12 (60) |
|
1 |
6 (30) |
|
2 |
2 (10) |
|
Palliative SEMS |
9 (45) |
|
Bridge to Surgery |
11 (55) |
|
Missing data |
|
|
1 hour follow-up |
3 (15) |
|
4 hour follow-up |
5 (25) |
ASA, American Society of Anesthesiologists. SEMS, Self-expanding metallic stent
3.1 Inflammatory response
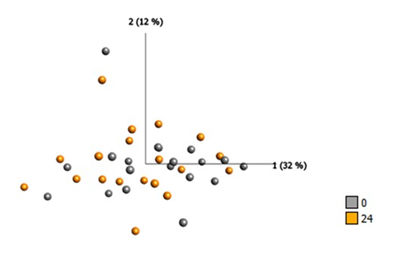
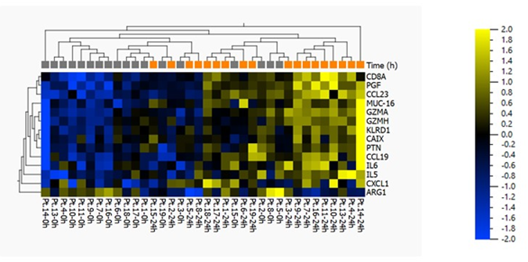
Protein quantification analysis targeting 92 predefined proteins was performed on plasma samples. Baseline samples were compared with samples collected 24 hours after SEMS placement. The fourteen proteins that differed the most between hours 0 and 24 are listed in Table 2. There was a significant difference in eleven protein concentrations (ARG1, GZMA, PGF, GZMH, KLRD1, CCL23, IL5, CCL19, PTN, MUC-16) and a tendency towards higher levels of three proteins (CAIX, CXCL1, IL6) (Figure 1a and 1b).
Figure 2(b): Heat map and two-way unsupervised hierarchical clustering based on the 14 proteins that differ the most between baseline and 24 hours after SEMS placement. This is a paired analysis (where the patient is eliminated as a factor, and subject #12 has been excluded from the analysis) with the following “loose” filtering criteria: SD>0.2, p<0.10, q=0.23, and >1.2-fold difference between the two groups. If standard criteria were to be applied, i.e. p<0.05 and >2-fold change, then no proteins would have passed this more stringent filter.
Table 2: Overview of proteins levels that differentiated the most after SEMS placement for malignant obstruction
|
Protein |
Difference |
P-value |
Immune Response |
Proliferation/Cell survival |
Angiogenesis |
CRC |
Tumerigenic |
Anti-tumor |
|
ARG1 |
-0.48 |
0.029 |
x |
x |
||||
|
GZMA |
0.27 |
0.012 |
x |
x |
x |
|||
|
PGF |
0.27 |
0.003 |
x |
x |
x |
|||
|
CD8A |
0.28 |
0.004 |
x |
x |
||||
|
CAIX |
0.3 |
0.092 |
x |
x |
x |
|||
|
GZMH |
0.31 |
0.015 |
x |
|||||
|
KLRD1 |
0.31 |
0.03 |
x |
x |
x |
|||
|
CCL23 |
0.32 |
0.017 |
x |
x |
||||
|
IL5 |
0.33 |
0.044 |
||||||
|
CCL19 |
0.34 |
0.039 |
x |
x |
||||
|
PTN |
0.37 |
0.041 |
x |
x |
||||
|
CXCL1 |
0.41 |
0.087 |
x |
x |
x |
|||
|
IL6 |
0.47 |
0.067 |
x |
x |
x |
x |
||
|
MUC-16 |
0.49 |
0.004 |
x |
x |
Difference is defined as log2-fold change
3.1.1 Tumorigenic inflammation
Significantly increased concentrations were found for granzyme A (GZMA) (log2-fc: 0.27, p-value = 0.012) and granzyme H (GZMH) (log2-fc: 0.31, p-value = 0.015). Granzymes, a family of serine proteases, are expressed exclusively by cytotoxic T lymphocytes and natural killer (NK) cells. In colorectal cancer, GZMA has been shown to induce tumor promoting inflammation [24], whereas, increased gene expression of GZMH has been associated with right-sided CRC, however, the biological function is unknown [25].
Killer cell lectin receptor D1 (KLRD1/CD94) (log2-fc: 0.31, p-value = 0.030) was significantly increased and have recently been suggested as an immune checkpoint in CRC and a potential target for immunotherapy [26]. Additionally, higher concentrations for interleukine 5 (IL5) (log2-fc: 0.33, p-value = 0.044) and mucin 16 (MUC-16) (log2-fc: 0.49, p-value = 0.048) were found. Interleukine 5 is an inflammatory marker and MUC-16, also known as CA-125, which is a biomarker used in ovarian cancer [27].
3.1.2 Angiogenesis
Significantly increased concentrations were found for placental growth factor (PGF) (log2-fc: 0.27, p-value = 0.003), a member of the vascular endothelial growth factor (VEGF) family [28], and pleiotrophin (PTN) (log2-fc: 0.37, p-value = 0.041), a cytokine that induces angiogenesis by promoting VEGF expression [29]. Additionally, significantly elevated concentration of chemokine ligand 23 (CCL23) (log2-fc: 0.32, p-value = 0.017) was seen, an inflammatory cytokine which have been associated with angiogenic properties in CRC [30].
3.1.3 Migration
Increased concentration of chemokine ligand 1 (CXCL1) (log2-fc: 0.41, p-value = 0.087), however, not statistically significant. Chemokine ligand 1 is involved with the migration of tumor cells and premetastatic niche formation in CRC [31].
3.1.4 Proliferation
A tendency towards higher protein levels found for carbonic anhydrase 9 (CAIX) (log2-fc: 0.31, p-value = 0.093) and interleukin 6 (IL6) (log2-fc: 0.47, p-value = 0.067). Both are known to promote proliferation and cell survival in CRC [32-36].
3.1.5 Anti-tumor function
Decreased protein concentration of Arginase 1 (ARG1) was found (log2-fold change (fc), -0.48, p-value = 0.029), an enzyme which have been associated with poor prognosis in CRC. Additionally, higher concentration of chemokine ligand 19 (CCL19) (log2-fc: 0.34, p-value = 0.034) which have suggested to suppress angiogenesis in colorectal cancer [37], and CD8A (log2-fc: 0.28, p-value = 0.004) which is a mediator of adaptive immunity, and important for killing cancerous or virally infected cells [38, 39].
3.2 Cell-free DNA (cfDNA)
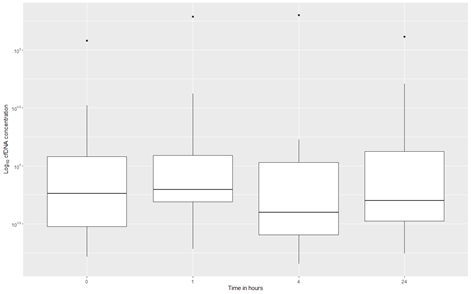
Quantitative analyses of circulating cell-free DNA (cfDNA) were performed at four different time points. Blood samples were collected at baseline and again 1, 4, and 24 hours after SEMS placement (Figure 2). Due to missing data, a total of twelve patients were included in the analysis. The circulating cfDNA concentration of the participants did not significantly change during the first 24 hours after stenting, , p= 0.3592.
Figure 2: Boxplots of the log10 transformed concentration of cell free DNA at 0, 1, 4 and 24 hours after stenting. The boxplots show the 1st quartile, median and 3rd quartile, the upper- and lower whiskers extend a maximum of 1.5 times the interquartile range from respectively the 3rd quartile to the largest value or the 1st quartile to the lowest value. Measurements outside this range are considered outlying measurements and are plotted individually as points. cfDNA: Cell free DNA
4. Discussion
This prospective study included 20 patients undergoing acute SEMS insertion for malignant colonic obstruction. This is the first study to investigate the systemic inflammatory response after SEMS placement. Protein quantification analysis was performed on plasma samples and found 11 proteins with significantly changed concentrations 24 hours after SEMS placement. In addition, quantitative analyses of circulating cfDNA were performed, and no significant differences were found between the time points 0, 1, 4, and 24 hours after SEMS placement.
It is well known that inflammation is closely associated with cancer development and progression in CRC [40-43]. In theory, SEMS placement induces inflammation due to tissue damage, ischemia, and necrosis which may be followed by acute and chronic inflammation. We investigated the systemic inflammatory response through a protein biomarker proteomics panel. Proteins represent the essential functionality in understanding disease pathology and associated biological processes, and several proteins in the biomarker panel are associated with essential hallmarks of cancer, including inflammation, proliferation, migration, and angiogenesis [40, 44]. Among the 14 proteins that differed the most between before and after SEMS placement, increased concentrations were found for thirteen proteins, of which eight (GZMA, PGF, CAIX, KLRD1, PTN, CXCL1, IL-6, MUC-16) have been associated with tumor-promoting functions in CRC (24,27,28,34,36,45-47). The impact of interleukine 5 (IL-5) on CRC is unclear, however, IL-5 is an inflammatory cytokine, that has been associated with tumor progression in breast cancer [48].
We found the highest increase for MUC-16, which is known as a biomarker in ovarian cancer [27]. MUC-16 allows the binding of cancer cells to mesothelin and has been reported as a protein crucial in the peritoneal dissemination of ovarian cancer through adhesion [49, 50]. Interestingly, MUC-16 has also been shown to have a negative impact on prognosis in CRC (27). Increased concentrations were found for three proteins (PGF, PTN, CCL23) that have been associated with angiogenic function (28-30), and PGF and PTN are both associated with VEGF expression [29]. Furthermore, PGF is activated in inflammation and associated with angiogenesis through WNT2 signaling in CRC [51]
In addition, granzyme A (GZMA) (log2-fc: 0.27, p-value = 0.012) was found to be increased after SEMS. Granzyme A is a pro inflammatory protease that promotes CRC development. Among several functions, GZMA stimulates IL-6 production in macrophages (24). An interesting find was a tendency toward a higher concentration of IL-6 after SEMS, which is known to be associated with acute and chronic inflammation and it is well documented that IL6 is associated with proliferation in CRC [34, 47, 52-54], and increases the risk of recurrence [34].
We found increased levels of CD8A, associated with anti-tumor effects [38] and CCL19, which have been shown to suppress angiogenesis in CRC [37]. When a systemic stress response is initiated, we usually expect to see a fall in CD8a concentration within hours. The increased concentration of CD8a was a surprising find. Maybe it is due to a low number of patients, or a neoantigen surge due to cell turnover. Additionally, the analysis found decreased levels of ARG1 after stent placement, a protein that is associated with a poor prognosis in CRC [55].
Our analysis primarily found increased levels of proteins associated with tumor progression, such as proliferation and angiogenesis [19-29, 33], which indicates that stent insertion may be initiating an adverse pro-metastatic systemic response. However, a limitation to the study is the small number of included patients, making the study explorative, and should be interpreted with caution. Furthermore, measurements were made immediately after the procedure and in a short period. Interesting changes in several protein levels may be missed. The Olink panel includes biomarkers related to a wide range of mechanisms, including the adaptive immune response and it is well known that an adaptive immune response can be detected after 4-7 days. The analysis found indication of an unfavorable response, however, no further follow-up was made, and whether these results translate into impaired prognosis is only speculative.
Previous evidence has shown elevated levels of circulating cfDNA after SEMS placement [12], which has been associated with poor prognosis in CRC [13, 14]. A study found significantly elevated levels on day 7; however, no difference was found on days 1 and 3 [12]. This correlates with our findings as no significant difference was found between baseline and 1, 4, and 24 hours after SEMS placement. These results could indicate that the cell damage may be caused by a persistent mechanical pressure rather than the acute effects of the SEMS procedure itself. However, our findings were limited by a relatively small number of patients and should be interpreted with caution.
In conclusion, the protein profile after stent placement does not unanimously reflect a pro-metastatic phenotype within the first 24 hours. However, several of the biomarkers investigated may be candidate outcome measures in future studies investigating stent-placement-related pharmacological blockade of inflammation, and this study supports the importance of further investigation in the systemic response following SEMS placement for obstructing CRC.
Author disclosure
The authors, Malene Broholm, Rasmus Bojesen, Mikail Gögenur, Andreas Weinberger Rosen, Sara Watt, Mustafa Bulut, Rasmus Vogelsang, Adile Orhan, Ellen Bjerrum, Christina Søs Auður Andersen, Niels Pallisgaard, Thomas Litman, and Ismail Gögenur have no conflicts of interest or financial ties to disclose
References
- Angell HK, Bruni D, Barrett JC, Herbst R & Galon J. The Immunoscore: Colon Cancer and Beyond. Clinical Cancer Research?: An Official Journal of the American Association for Cancer Research 26 (2020): 332–339.
- Aquina CT, Becerra AZ, Xu Z, Boscoe FP, Schymura MJ, Noyes K, et al. Nonelective colon cancer resection: A continued public health concern. Surgery 161 (2017): 1609–1618.
- Assarsson E, Lundberg M, Holmquist G, Björkesten J, Thorsen SB, Ekman D, et al. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability PloS One 9 (2014): e95192.
- Baron TH & Harewood GC. Enteral self-expandable stents. Gastrointestinal Endoscopy 58 (2003): 421–433.
- Benjamini Y & Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Stat. Methodol 57 (1995): 289–300.
- Bioxpedia (nd). https://www.bioxpedia.com/olink-proteomics/
- Björkman K, Mustonen, H, Kaprio T, Haglund C & Böckelman C. Mucin 16 and kallikrein 13 as potential prognostic factors in colon cancer: Results of an oncological 92-multiplex immunoassay. Tumour Biology?: The Journal of the International Society for Oncodevelopmental Biology and Medicine 41 (2019): 1010428319860728.
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA & Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians 68 (2018): 394–424.
- Coelho R, Marcos-Silva L, Ricardo S, Ponte F, Costa A, Lopes JM, et al. Peritoneal dissemination of ovarian cancer: Role of MUC16-mesothelin interaction and implications for treatment. Expert Review of Anticancer Therapy 18 (2018): 177–186.
- Ducoin K, Oger R, Mutala LB, Deleine C, Jouand N, Desfrançois J, et al. Targeting NKG2A to boost anti-tumor CD8 T-cell responses in human colorectal cancer (2022).
- Escudero-Esparza A, Martin TA, Davies ML & Jiang WG. PGF isoforms, PLGF-1 and PGF-2, in colorectal cancer and the prognostic significance. Cancer Genomics & Proteomics 6 (2009): 239–246.
- Eugène J, Jouand N, Ducoin K, Dansette D, Oger R, Deleine C, et al. The inhibitory receptor CD94/NKG2A on CD8 (+) tumor-infiltrating lymphocytes in colorectal cancer: a promising new druggable immune checkpoint in the context of HLAE/β2m overexpression. Modern Pathology?: An Official Journal of the United States and Canadian Academy of Pathology Inc 33 (2020): 468–482.
- Fernández-Esparrach G, Bordas JM, Giráldez MD, Ginès A, Pellisé, M, Sendino O, et al. Severe complications limit long-term clinical success of self-expanding metal stents in patients with obstructive colorectal cancer. The American Journal of Gastroenterology 105 (2010): 1087–1093.
- Foo CC, Poon SHT, Chiu RHY, Lam WY, Cheung LC & Law WL. Is bridge to surgery stenting a safe alternative to emergency surgery in malignant colonic obstruction: a meta-analysis of randomized control trials. Surgical Endoscopy 33 (2019): 293–302.
- Galon J, Mlecnik B, Bindea G, Angell HK, Berger A, Lagorce C, et al. Towards the introduction of the “Immunoscore” in the classification of malignant tumours. The Journal of Pathology 232 (2014): 199–209.
- Greten FR & Grivennikov SI. Inflammation and Cancer: Triggers, Mechanisms, and Consequences. Immunity 51 (2019): 27–41.
- Grivennikov SI, Greten FR & Karin M. Immunity, inflammation, and cancer Cell 140 (2010): 883–899.
- Gubbels JAA, Belisle J, Onda M, Rancourt C, Migneault M, Ho M, Bera TK, et al. Mesothelin-MUC16 binding is a high affinity, N-glycan dependent interaction that facilitates peritoneal metastasis of ovarian tumors. Molecular Cancer 5 (2006).
- Hanahan D & Weinberg RA. Hallmarks of cancer: the next generation. Cell 144 (2011): 646–674.
- Heichler C, Scheibe K, Schmied A, Geppert CI, Schmid B, Wirtz S, et al. STAT3 activation through IL-6/IL-11 in cancer-associated fibroblasts promotes colorectal tumour development and correlates with poor prognosis Gut 69 (2020): 1269–1282.
- Iversen LH, Bülow S, Christensen IJ, Laurberg S & Harling H. Postoperative medical complications are the main cause of early death after emergency surgery for colonic cancer. The British Journal of Surgery 95 (2008), 1012–1019.
- Kim BG, Gao M-Q, Kang S, Choi YP, Lee JH, Kim JE, et al. Mechanical compression induces VEGFA overexpression in breast cancer via DNMT3A-dependent miR-9 downregulation. Cell Death & Disease 8 (2017): e2646.
- Kong Y, Bai PS, Nan KJ, Sun H, Chen NZ & Qi XG. Pleiotrophin is a potential colorectal cancer prognostic factor that promotes VEGF expression and induces angiogenesis in colorectal cancer. International Journal of Colorectal Disease 27 (2012): 287–298.
- Liang L, Zeng J-H, Qin X-G, Chen J-Q, Luo D-Z & Chen G. Cellular Physiology and Biochemistry Cellular Physiology and Biochemistry Original Paper Distinguishable Prognostic Signatures of Left-and Right-Sided Colon Cancer: a Study Based on Sequencing Data Cellular Physiology and Biochemistry Cellular Physiology and Biochemistry. Cell Physiol Biochem 48 (2018): 475–490.
- Ma Z, Lian J, Yang M, Wuyang J, Zhao C, Chen W, et al. Overexpression of Arginase-1 is an indicator of poor prognosis in patients with colorectal cancer. Pathology, Research and Practice 215 (2019): 152383.
- McLean MH, Murray GI, Stewart KN, Norrie G, Mayer C, Hold GL. The inflammatory microenvironment in colorectal neoplasia. PloS One 6 (2011): e15366.
- Olink proteomics. https://www.bioxpedia.com/wp-content/uploads/2020/04/1047-v1.2-Immuno-Onc-Panel-Content_final.pdf (2019).
- Olsen J, Kirkeby LT, Olsen J, Eiholm S, Jess P, Gögenur I & Troelsen JT. High interleukin-6 mRNA expression is a predictor of relapse in colon cancer. Anticancer Research 35 (2015): 2235–2240.
- Pallisgaard N, Spindler K-LG, Andersen RF, Brandslund I & Jakobsen A. Controls to validate plasma samples for cell free DNA quantification. Clinica Chimica Acta; International Journal of Clinical Chemistry 446 (2015): 141–146.
- Pucciarelli S, Zorzi M, Gennaro N, Gagliardi G, Restivo A, Saugo M, et al. In-hospital mortality, 30-day readmission, and length of hospital stay after surgery for primary colorectal cancer: A national population-based study. European Journal of Surgical Oncology?: The Journal of the European Society of Surgical Oncology and the British Association of Surgical Oncology 43 (2017): 1312–1323.
- Quail DF, Olson OC, Bhardwaj P, Walsh LA, Akkari L, Quick ML, et al. Obesity alters the lung myeloid cell landscape to enhance breast cancer metastasis through IL5 and GM-CSF. Nature Cell Biology 19 (2017): 974–987.
- Ribeiro IB, de Moura DTH, Thompson CC & de Moura EGH. Acute abdominal obstruction: Colon stent or emergency surgery? An evidence-based review. World Journal of Gastrointestinal Endoscopy 11 (2019): 193–208.
- Santer FR, Malinowska K, Culig Z & Cavarretta IT. Interleukin-6 trans-signalling differentially regulates proliferation, migration, adhesion and maspin expression in human prostate cancer cells. Endocrine-Related Cancer 17 (2010): 241–253.
- Santiago L, Castro M, Sanz-Pamplona R, Garzón M, Ramirez-Labrada A, Tapia E, et al. Extracellular Granzyme A Promotes Colorectal Cancer Development by Enhancing Gut Inflammation. Cell Reports 32 (2020): 107847.
- Sjo OH, Larsen S, Lunde OC & Nesbakken A. Short term outcome after emergency and elective surgery for colon cancer. Colorectal Disease?: The Official Journal of the Association of Coloproctology of Great Britain and Ireland 11 (2009): 733–739.
- Takahashi G, Yamada T, Iwai T, Takeda K, Koizumi M, Shinji S et al. Oncological Assessment of Stent Placement for Obstructive Colorectal Cancer from Circulating Cell-Free DNA and Circulating Tumor DNA Dynamics. Annals of Surgical Oncology 25 (2018): 737–744.
- Tanaka T, Narazaki M & Kishimoto T. IL-6 in inflammation, immunity, and disease. Cold Spring Harbor Perspectives in Biology 6 (2014): a016295.
- Terzi? J, Grivennikov S, Karin E & Karin M. Inflammation and colon cancer. Gastroenterology 138 (2010): 2101-2114.
- Tse JM, Cheng G, Tyrrell JA, Wilcox-Adelman SA, Boucher Y, Jain RK. Mechanical compression drives cancer cells toward invasive phenotype. Proceedings of the National Academy of Sciences of the United States of America 109 (2012): 911–916.
- Tülüce Y, Ahmed BA, Koyuncu ? & Durgun M. The cytotoxic, apoptotic and oxidative effects of carbonic anhydrase IX inhibitor on colorectal cancer cells. Journal of Bioenergetics and Biomembranes 50 (2018): 107–116.
- Unterleuthner D, Neuhold P, Schwarz K, Janker L, Neuditschko B, Nivarthi H et al. Cancer-associated fibroblast-derived WNT2 increases tumor angiogenesis in colon cancer. Angiogenesis, 23 (2020): 159–177.
- van der Bij GJ, Oosterling SJ, Beelen RHJ, Meijer S, Coffey JC & van Egmond M. The perioperative period is an underutilized window of therapeutic opportunity in patients with colorectal cancer. Annals of Surgery 249 (2009): 727–734.
- Van Hooft JE, Veld Jv, Arnold D, Beets-Tan RGH, Everett S, Götz M, et al. Self-expandable metal stents for obstructing colonic and extracolonic cancer: European Society of Gastrointestinal Endoscopy (ESGE) Guideline - Update 2020. Endoscopy 52 (2020): 389–407.
- Vymetalkova V, Cervena K, Bartu L & Vodicka P. Circulating Cell-Free DNA and Colorectal Cancer: A Systematic Review. International Journal of Molecular Sciences 19 (2018).
- Waldner MJ, Foersch S & Neurath MF. Interleukin-6--a key regulator of colorectal cancer development. International Journal of Biological Sciences 8 (2012): 1248–1253.
- Wang D, Sun H, Wei J, Cen B & DuBois RN. CXCL1 Is Critical for Premetastatic Niche Formation and Metastasis in Colorectal Cancer. Cancer Research 77 (2017a): 3655–3665.
- Wang D, Sun H, Wei J, Cen B & DuBois RN. CXCL1 Is Critical for Premetastatic Niche Formation and Metastasis in Colorectal Cancer. Cancer Research 77 (2017b): 3655–3665.
- Wang T, Song P, Zhong T, Wang X, Xiang X, Liu Q, et al. The inflammatory cytokine IL-6 induces FRA1 deacetylation promoting colorectal cancer stem-like properties. Oncogene 38 (2019): 4932–4947.
- Wang X, Shi X-Q, Zeng P-W, Mo F-M & Chen Z-H. Circulating cell free DNA as the diagnostic marker for colorectal cancer: a systematic review and meta-analysis. Oncotarget 9 (2018): 24514–24524.
- Watt SK, Hasselbalch HC, Skov V, Kjær L, Thomassen M, Kruse TA, et al. Whole Blood Gene Expression Profiling in patients undergoing colon cancer surgery identifies differential expression of genes involved in immune surveillance, inflammation and carcinogenesis. Surgical Oncology 27 (2018): 208–215.
- Watt SK, Hasselbalch HC, Skov V, Kjær L, Thomassen M, Kruse TA, et al. Increased oxidative stress with substantial dysregulation of genes related to oxidative stress and DNA repair after laparoscopic colon cancer surgery. Surgical Oncology 35 (2020): 71–78.
- Xu C, Zhu S, Wu M, Han W & Yu Y. Functional receptors and intracellular signal pathways of midkine (MK) and pleiotrophin (PTN). Biological & Pharmaceutical Bulletin 37 (2014): 511–520.
- Xu J, Ye Y, Zhang H, Szmitkowski M, Mäkinen MJ, Li P, et al. Diagnostic and Prognostic Value of Serum Interleukin-6 in Colorectal Cancer. Medicine 95 (2016): e2502.
- Xu Z, Zhu C, Chen C, Zong Y, Feng H, Liu D, et al. CCL19 suppresses angiogenesis through promoting miR-206 and inhibiting Met/ERK/Elk-1/HIF-1α/VEGF-A pathway in colorectal cancer. Cell Death & Disease 9 (2018): 974.
- Yang P, Lin X-F, Lin K & Li W. The Role of Stents as Bridge to Surgery for Acute Left-Sided Obstructive Colorectal Cancer: Meta-Analysis of Randomized Controlled Trials. Revista de Investigacion Clinica; Organo Del Hospital de Enfermedades de La Nutricion 70 (2018): 269–278.
- Ying J, Tsujii M, Kondo J, Hayashi Y, Kato M, Akasaka T et al. The effectiveness of an anti-human IL-6 receptor monoclonal antibody combined with chemotherapy to target colon cancer stem-like cells. International Journal of Oncology 46 (2015): 1551–1559.


 Impact Factor: * 3.5
Impact Factor: * 3.5 Acceptance Rate: 71.36%
Acceptance Rate: 71.36%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 
