Molecular Docking and ADMET Prediction of Modified and Derivative forms of 2-Deoxy-D-glucose Against COVID-19 main Protease Complex: In silico Approach
Rashmi Rekha Samal 1, 2, Suryasikha Samal3, Choudhury Suryakant Mishra4, Atala Bihari Jena5 and Asim K. Duttaroy*, 6
1CSIR-Institute of Minerals & Materials Technology, Bhubaneswar 751 013, India
2Academy of Scientific & Innovative Research (AcSIR), New Delhi 110025, India
3Department of Agriculture and Allied Sciences, C.V Raman Global University, India
4Odisha University of Agriculture and Technology, College of Basic Science and Humanities, India
5Complement Biology Laboratory, National Centre for Cell Science, S. P. Pune University campus, Pune 411007, India.
6Department of Nutrition, Institute of Basic Medical Sciences, Faculty of Medicine, University of Oslo, Norway
*Corresponding author: Asim K. Duttaroy, Department of Nutrition, Institute of Basic Medical Sciences, Faculty of Medicine, University of Oslo, Oslo, Norway.
Received: 11 February 2024; Accepted: 04 March 2024; Published: 20 March 2024
Article Information
Citation:
Rashmi Rekha Samal, Suryasikha Samal, Choudhury Suryakant Mishra, Atala Bihari Jena and Asim K. Duttaroy. Molecular Docking and ADMET Prediction of Modified and Derivative forms of 2-Deoxy-D-glucose Against COVID-19 main Protease Complex: In silico Approach. Fortune Journal of Health Sciences. 7 (2024): 138-150.
View / Download Pdf Share at FacebookAbstract
The emergence of the Coronavirus outbreak presented a worldwide challenge, demanding effective therapeutic strategies with minimal toxicity. In this regard, potential drug candidates are being explored. One such candidate is 2-Deoxy-D-glucose, prescribed by India's Defense Research and Development Organization (DRDO). To determine its effectiveness against COVID-19, the derivative and modified form of 2-Deoxy-D-glucose were tested through in silico analysis against the COVID-19 main protease complex with an N3 inhibitor. The modified form of 2-Deoxy-D-glucose was created by replacing the hydroxy group with a hydrogen atom, and a derivative named Cypate 2-Deoxy-D-glucose was chosen to test against the COVID-19 main protease complex. The toxicity of these compounds was also scrutinized through ADMET analysis to predict their potential candidacy. Our data suggest that the modified version of 2-Deoxy-D-glucose is more stable and has minimal toxicity against the COVID-19 main protease, making it a possible candidate for further exploration.
Keywords
<p>Covid-19; Deoxy-D-glucose; molecular docking; ADMET</p>
Article Details
Introduction
SARS-CoV-2, the pathogen responsible for COVID-19, elicits a range of clinical complexities such as inflammation, cytokine storm, and multi-organ dysfunction. This outbreak has gained worldwide attention due to its significant impact [1]. As of February 2021, the global death toll had reached an estimated 2.3 million [2]. Nevertheless, controlled and therapeutic strategies are essential requisites to deal with the rising cases of infections. Before this pandemic, a group of researchers working at the Institute of Nuclear Medicine and Allied Sciences under the Defense Research and Development Organisation (DRDO) India developed a 2-Deoxy-D-glucose drug with the help of a pharmaceutical group to treat patients with moderate and severe COVID-19 symptoms. The past studies examined critically affected COVID patients by implementing Low Dose Radiation Therapy (LDRT) as a plausible method in pneumonia conditions [3]. COVID-19 patients receive treatment according to the severity of their disease. In India, oxygen support and intravenous steroids such as remdesivir and tocilizumab are administered to patients in the moderate to the severe category but restricted to selected patients. In May 2020, during the first wave of the pandemic, INMAS-DRDO scientists initiated a Phase II clinical trial on 2-Deoxy-D-glucose in COVID-19 patients. A Phase III study followed this. Finally, on May 1, 2021, the Drugs Controller General of India (DCGI) approved the emergency use of 2-Deoxy-D-glucose as an adjunct therapy in patients with moderate to severe COVID-19.
According to the interim data, the Phase-II trial reported a median time difference of 2.5 days to achieve normalization of specific vital signs parameters in the 2-Deoxy-D-glucose arm compared to Standard of Care. The use of therapeutic strategies in COVID-19 has been limited by the evolving side effects of antibodies and anti-viral agents like remdesivir, lopinavir, and other drugs, including heparin and atazanavir which have been investigated during drug-protein and protein-protein interactions [4,5]. The possibility of representing 2-Deoxy-D-glucose, previously designed for cancer, arose due to a potential antiviral drug against SARS-CoV-2. The influential role of 2-Deoxy-D-glucose was evident as a conjugate named Hyaluronic acid-2-Deoxy-D-Glucose against SARS-CoV-2 viral protein with better biodegradability and biocompatibility and minimal toxicity than 2-Deoxy-D-Glucose [6]. This analog of glucose plays a crucial role in the glycolytic pathway and leads to the inhibition of glycolysis.
Nonetheless, the adversity of 2-Deoxy-D-Glucose was conquered by combining with natural polymers, metal complexes, and azido analog, which was able to produce catastrophic oxidative stress in acute conditions of critically affected COVID-19 patients [7,8]. The effectiveness of 2-Deoxy-D-Glucose as a dietary component was previously scrutinized in vitro and in vivo by exploring its radio and chemo-sensitizing effects [9]. Nonetheless, the role of 2-Deoxy-D-Glucose in glycolysis acts as a competitive antagonist and inhibits viral replication and tumour cell proliferation [10]. Since the original 2-Deoxy-D-Glucose was generated from d-glucose by eliminating the hydroxyl group at the C-2 position [11]; the current study aimed to explore the potentiality of modified 2-Deoxy-D-Glucose against COVID-19. Previously, the removal of the hydroxyl group was compensated by hydrogen atoms and led to the inhibition of glycolysis, followed by the prevention of glucose-6- phosphate production [12]. The current study aims to identify a potential compound similar to 2-Deoxy-D-Glucose with less toxicity. In this regard, different derivatives are also considered for the comparative study. Among the derivatives of 2-Deoxy-D-Glucose, Cypate 2-Deoxy-D-Glucose was chosen as a COVID-19 inhibitor due to its optical fluorescence imaging ability. Cypate is a reactive carbocyanine and lipophilic dye where the evaluation of the compound was reported in tumor-bearing mice in vivo conditions through NIR optical imaging [13,14].
This study tested the potentiality of 2-Deoxy-D-Glucose molecules against COVID-A19 main protease complex with N3 inhibitor (PDB ID: 6LU7) in silico model. The main target protease (Mpro) of SARS-CoV-2: Mpro emerged as a critical enzyme of coronavirus and has a crucial role in regulating viral replication and transcription [15]. DockThor is an automated web server based on a global docking method utilized for flexible docking [16]. Furthermore, the potential candidate among the modified and derivative forms of 2-Deoxy-D-Glucose against COVID-19 main protease complex was identified based on minimal toxicity compared to the parent compound 2-Deoxy-D-Glucose.
Materials and Methods
The target PDB structure selection and validation
The target protein for this investigation was the crystal structure of COVID-19 main protease in complex with an inhibitor N3 with PDB ID: 6LU7. The structure with N3 inhibitor (2.16 Å resolutions) was downloaded from the RCSB database (https://www.rcsb.org/search). The selection was based on the importance of the Main protease (Mpro) of SARS-CoV-2 in mediating viral replication and transcription having close homologous with humans. After the protein retrieval from the database, the protein's validity was evaluated using SAVESv6.0 - Structure Validation Server. The overall quality factor was determined by ERRAT tool of SAVES v6.0 to recognize the correct regions of protein structures based on atomic interaction. Further, the overall G factor was determined by PROCHECK tool to analyze the quality of the selected target protein model. The degrees of unusualness and normality were predicted by G-factor based on stereochemical properties.
Active site prediction
The active site prediction of COVID-19 main protease was examined by CASTp 3.0 server using a Computational Geometry algorithm with basic ingredients such as Delaunay triangulation, alpha shape, and discrete flow (http://sts.bioe.uic.edu/castp/index.html). The area and the volume of the active site occupied by the residues were determined.
Ligand files preparation and optimization
The 3D structure of 2-Deoxy-Glucose (C6H12O5) and its derivative Cypate-2-Deoxy-D-glucose (C53H63N4O12) treated as candidate ligands against the target protein COVID-19 main protease. The structures were retrieved from the PubChem database in SDF format and converted to PDB format using BIOVIA Discovery Studio (https://pubchem.ncbi.nlm.nih.gov/). The modified 2-Deoxy-Glucose was prepared in Argus lab (4.0.1) by replacing two hydroxyl groups with polar hydrogen molecules. The structure was optimized by performing energy minimization using the UFF force field. The energy-minimized ligands were further analyzed in molecular docking experiments.
Molecular Docking
DockThor, a web tool for ligand-protein docking, was implemented to perform the molecular docking between the modified and derivative of 2-Deoxy-Glucose against COVID-19 main protease; both the receptor and ligands were optimized by applying the same force field known as MMFF94 Force Field throughout the docking experiments. DockThor program implemented the phenotypic crowding-based multiple solution steady-state genetic algorithms as the search method during molecular docking. As the blind docking was performed, the grid box parameter was set automatically by setting the centers X, Y, and Z as 22 Å. The complex's total energy (Etotal) was calculated based on intermolecular and intramolecular interaction energy by summing up the Van der Waals and electrostatic potentials between the protein-ligand atom pairs. All docking poses were generated during the docking step by utilizing the standard algorithm and then clustered with the help of the DTStatistics tool. The number of docking runs was set by 24; for each docking run, maximum 20 numbers of clusters were generated. The top energy poses of each cluster were screened as best reprehensive of 2-Deoxy-Glucose ligands against COVID-19 main protease.
Prediction of ADMET properties
An approaching compound's toxicity and clinical safety concerns are significant problems in drug discovery. Hence, the pharmacokinetics and toxicity of candidate ligands against COVID-19 main protease were screened by ADMETlab 2.0: an integrated online platform that helps evaluate accurate and comprehensive ADMET properties. The SMILES chemical structures of the parent, modified, and derivative of 2-Deoxy-Glucose were submitted to the server. The complete ADMET profiles of ligands were created after the generation of the model framework. The ligands described here were subjected to ADMET prediction to investigate the physiochemical properties of these compounds during the interaction with COVID-19 main protease. For the binding purpose, physicochemical descriptors with vital roles were selected, including molecular weight, logP, logS, hydrogen acceptor and donor, polar surface area, F30% human oral bioavailability, and Human intestinal absorption (HIA). These are the critical and essential prerequisites to determine its apparent efficacy and related to oral bioavailability. Plasma protein binding (PPB) is an essential mechanism of drug uptake and distribution by binding to plasma proteins indicating a powerful influence on pharmacodynamic behavior. Other properties, such as drug clearance (CL) and half-life (T1/2), are essential pharmacokinetic parameters to study the influence on drugs. Drug-induced liver injury (DILI) and mutagenicity are vital descriptors to evaluate the safety of medicines. These descriptors helped correlate the binding pose and binding energy formed during the formation of the COVID-19 main protease complex with 2-Deoxy-Glucose molecules. ADMET analysis provided a better mechanistic understanding of binding relationships with COVID-19 main protease. It determined the suitable ligand candidate for implementation as the essential medication and strategy to treat a viral infection like COVID in this pandemic.
Results and Discussion
Structural evaluation and optimization of COVID-19 main protease complex
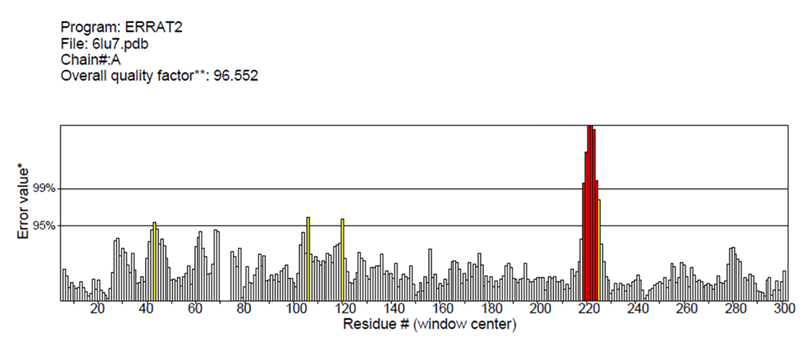
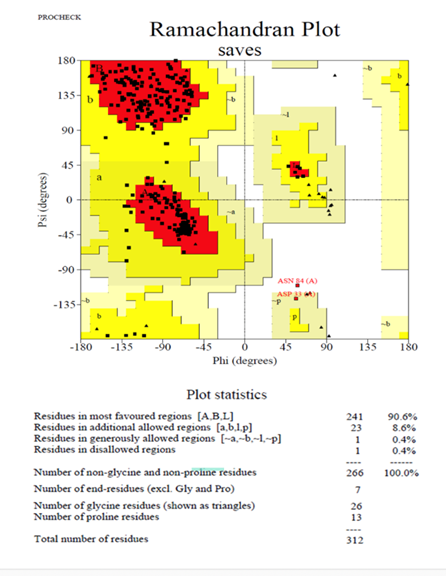
The structure of COVID-19 main protease in complex with an inhibitor N3 (PDB 6LU7), formerly known as SARS-CoV-2 Mpro, plays a vital role in regulating viral replication and transcription, which could be used as a drug target to elucidate the binding mechanism. [17,18]. Developing a single antiviral agent targeting Mpro, or combining such an agent with other potential therapies, could provide an effective first line of defense against all coronavirus-related diseases. Due to the functional importance of Mpro in the viral life cycle, it can be implemented as a potential target for developing antiviral drugs after the structural evaluation and submission of the 3D structure to the RCSB database [15]. Although the enzymatic activity of COVID-19 main protease in Vero cells was measured against potential inhibitors such as N3 and Ebselen [15], in silico model evaluation is still to be explored. Ebselen and N3 inhibited SARS-CoV-2 with half-maximal effective concentration (EC50) values of 4.67 µM and 16.77 µM, respectively. Both of these compounds may penetrate the cellular membrane to reach their targets. The structural validation of the COVID-19 main protease complex is an essential prerequisite before predicting the binding mechanism with 2-Deoxy-Glucose molecules. For this purpose, the ERRAT tool of SAVES v6.0, an online server, was implemented to approve the protein structure based on the overall quality factor, contributing to non-bonded atomic interactions. Within proteins, atoms are distributed non-randomly concerning one another, influenced by complex geometric and energetic considerations [19]. The exceptional was maintained by excluding the residues covalently bonded to each other. The higher score symbolizes the high quality of the structure. The basic acceptance rate should be >50 to establish a high-quality model [20]. However, the overall quality factor of COVID-19 main protease was found to be 99.552, which was within a highly acceptable range (Figure 1). Furthermore, the Ramachandran plot of COVID-19 main protease was evaluated using PROCHECK software (Figure 2).
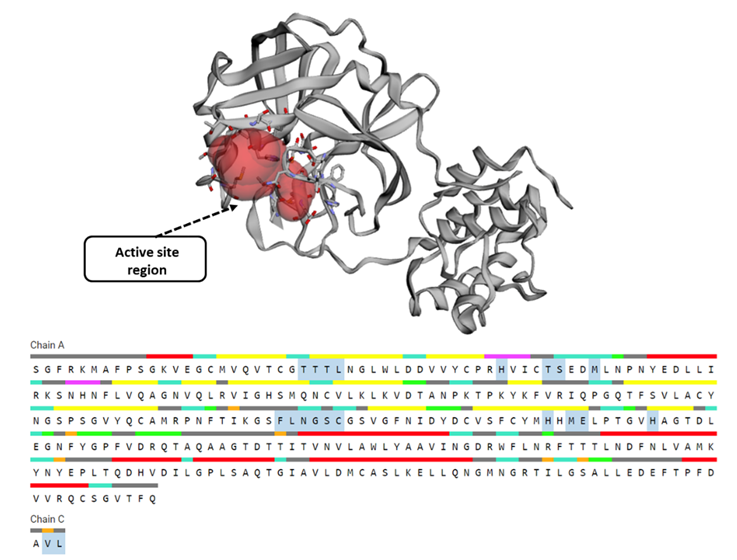
The residues in the most favorable region were 90.6%, while 8.6% were in the allowed region and only 0.4% in the disallowed region, confirming the target protein as a good quality model to perform the docking. PROCHECK also predicts the structure quality based on G-factors, which measure the unusualness [21]. Recently, researchers validated the spike glycoprotein through PROCHECK to obtain immunogenic epitopes [22]. Hence, by correlating the ERRAT value, the model had a 0.12 G-factor value significantly lower than the expected exceptional value (0.5), indicating the model's efficacy. Subsequently, the active site prediction of the COVID-19 main protease complex was performed by CASTp 3.0 server and discovered the amino acid residues in the active site cavity. Polar uncharged amino acids like Thr dominated the active site of COVID-19's main protease. Followed by Thr, the cavity contained the polar positively charged amino acid His and other amino acids such as Ser, Leu, and Met. Other residues like Phe140, Asn142, Gly143, Cys145, and Glu166 were present in the tunnel of protein (Figure 3).
Figure 3: Active site of COVID-19 main protease complex with N3 inhibitor predicted by CASTp 3.0 server. The area in red represents the active site of a protein and residues were represented in ball and stick manner. The sequence of COVID-19 main protease was presented by CASTp, where the highlighted residues in both chain A and C represents the active amino acid residues.
Earlier, Hatada and his group analysed the key residues during complex formation with inhibitors through fragment molecular orbital study and reported that His41, His163, His164, and Glu166 were the critical residues [23]. The active site residues have mouth openings that connect their interior to the outside bulk solution by developing concave surfaces, which aids in presenting the amino acid residues in a suitable configuration for the drug compound binding [24]. Hence, these residues of the COVID-19 main protease complex constructively influence the facilitation and adaptation of binding pockets to regulate binding processes and specific functionalities.
Molecular docking of COVID-19 main protease complex with a modified and derivative form of 2-Deoxy-Glucose
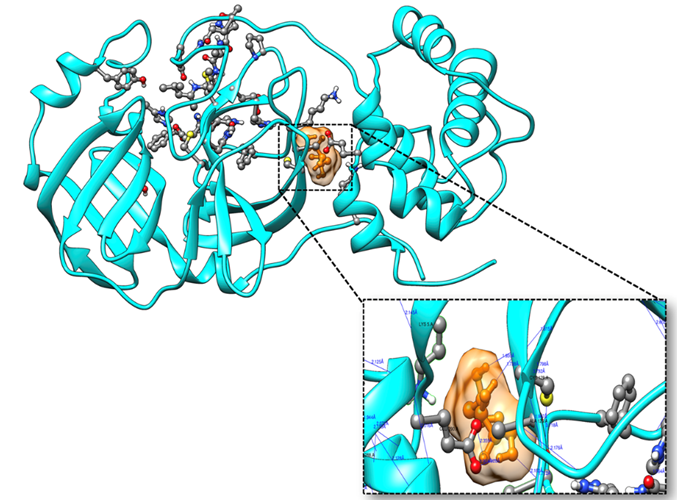
The optimized and validated COVID-19 main protease complex structure by SAVES v6.0 was exposed to molecular docking to uncover the binding mechanism against DRDO's prescribed anti-covid potential drug 2-Deoxy-Glucose, along with its modified and derivative forms. The results obtained from this binding analysis could help in establishing the opening of new research avenues. By performing the docking study of the COVID-19 main protease complex with 2-Deoxy-Glucose ligands using the DockThor, the top pose with the lowest binding energy and affinity was selected (Figure 4).
Figure 4: Molecular docking of the crystal structure of COVID-19 main protease complex (PDB ID:6LU7) with 2-Desoxy-D-glucose. The COVID-19 main protease complex was shown in cyan color in ribbon form, and 2-Desoxy-D-glucose was represented in a ball and stick model in the orange color surface. The enlarged version shows the binding mode of top-ranked docked poses and interacting important residues of COVID-19 main protease with 2-Desoxy-D-glucose by forming hydrogen bonds. The docking was performed in AutoDock 4.2, and visualization of the complex was done by UCSF Chimera software.
The docking station, DockThor, was used to predict the specific binding of ligands to the receptor using empirical scoring functions [25]. The comparative analysis among different forms of 2-Deoxy-Glucose with target protein was done to identify the competent compound against COVID viral infection. Initially, the parental compound, 2-Deoxy-Glucose retrieved from the PubChem database, was examined through docking, where the binding affinity was found to be -5.68 kcal/mol (Table 1). The Vander Waal and electrostatic energy were estimated by 0.76 and -34.78 kcal/mol, respectively. The binding energy of the complex formed by COVID-19 main protease and 2-Deoxy-Glucose was determined by -2.065 kcal/mol. The binding mechanism explored the critical amino acid residues that were participating in forming a complex with 2-Deoxy-Glucose. Mainly, Gln127, Ala129, Lys137, and Glu290 were found to be involved in forming hydrogen bonds with the oxygen atom of 2-Deoxy-Glucose at O1, O2, and O4 positions (Table 2).
Table 1: Binding parameters of the complex formed between COVID-19 main protease and ligands in DockThor.
|
Ligands |
Affinity (kcal/mol) |
VDW energy (kcal/mol) |
Electrostatic energy (kcal/mol) |
Binding energy (kcal/mol) |
|
2-Deoxy-D-glucose |
-5.683 |
0.76 |
-34.78 |
-2.065 |
|
Modified 2-Deoxy-D-glucose |
-5.821 |
-2.622 |
-19.69 |
-39.263 |
|
Cypate 2-Deoxy-D-glucose |
-9.18 |
-29.34 |
-25.43 |
-8.01 |
Table 2: Hydrogen bonds formed between COVID-19 main protease complex and atoms of 2-Deoxy-D-glucose, modified 2-Deoxy-D-glucose and cypate
|
Ligands |
Atom name of ligands |
Residue name of receptor |
Distance in Å |
|
2-Deoxy-D-glucose |
O2 |
Gln127 |
2.9 |
|
O1 |
Gln127 |
2.6 |
|
|
O4 |
Ala129 |
3.11 |
|
|
O4 |
Lys137 |
3.13 |
|
|
O1 |
Glu290 |
2.6 |
|
|
O4 |
Glu290 |
2.9 |
|
|
Modified 2-Deoxy-D-glucose |
O7 |
Lys5 |
2.75 |
|
O8 |
Cys128 |
3.06 |
|
|
O8 |
Ala129 |
3.24 |
|
|
O8 |
Lys137 |
3.02 |
|
|
O7 |
Glu290 |
2.76 |
|
|
Cypate 2-Deoxy-D-glucose |
O7 |
Lys5 |
3.13 |
|
N16 |
Gln110 |
2.86 |
|
|
O4 |
Thr111 |
1.43 |
|
|
O3 |
Cys128 |
3.2 |
|
|
O3 |
Ala129 |
2.54 |
|
|
O5 |
Lys137 |
2.25 |
|
|
O4 |
Asn151 |
1.89 |
|
|
O6 |
Cys160 |
1.48 |
|
|
N15 |
Glu290 |
2.03 |
|
|
O3 |
Glu290 |
2.66 |
|
|
O7 |
Glu290 |
2.42 |
However, the specific amino acid residues, such as Lys5 and Cys128, assisted in fabricating the hydrophobic region around the complex. Gln127 and Glu290 amplified the stability of the complex by forming the hydrogen bond with a 2.6 Å distance. In addition to the parental compound 2-Deoxy-Glucose, the modification was made to investigate the compound's efficacy against COVID-19 main protease by removing hydroxyl groups and replacing them with hydrogen atoms. Interestingly, the binding mechanism was indistinguishable to some extent from the parent compound, 2-Deoxy-Glucose (Figure 5). The residues such as Ala129, Lys137, and Glu290 were also found in common. Nevertheless, the bond length and the compound's orientation were changed by altering the position of the oxygen atom. Apart from these residues, Lys5 and Cys128 were found to be involved in complex formation by establishing the bond with 2.75 and 3.06 Å, respectively (Table 2). Tyr126 and Gln127 generated the hydrophobic region around the complex. And the Vander Waal and electrostatic energy were measured by -2.62 and -19.69 kcal/mol (Table 1).
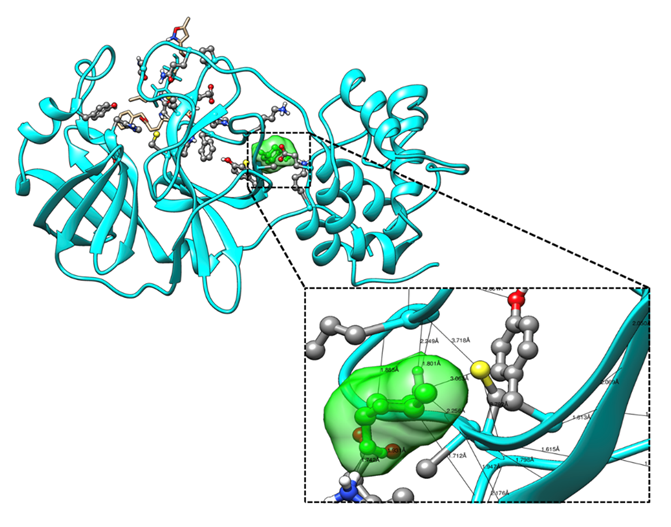
Figure 5: Molecular docking of the crystal structure of COVID-19 main protease complex (PDB ID:6LU7) with modified 2-Desoxy-D-glucose by removing two hydroxyl group. The COVID-19 main protease complex was shown in cyan color in ribbon form and modified 2-Desoxy-D-glucose was represented in ball and stick model in green color surface. The enlarged version showing the binding mode of top ranked docked poses and interacting important residues of COVID-19 main protease with modified 2-Desoxy-D-glucose by forming hydrogen bonds. The docking was performed in AutoDock 4.2 and visualization of the complex was done by UCSF Chimera software.
On the other hand, the binding energy was exceptionally distinctive from the parent compound, i.e., -39.26 kcal/mol. It contributed the maximum to the stability of the complex. The binding energy or free energy evaluation was a key factor that helped identify critical residues against potential inhibitors' design [26]. The reported studies on COVID-19 main protease described the binding mechanism with few natural, synthetic compounds such as flavonoids, peptides, terpenes, and quinolines. These complex formations were dominated by hydrogen and hydrophobic interaction involving key residues such as Gln89, Asn147, Glu166, and Gln189 [27,28]. Subsequently, the derivative form, Cypate 2-Deoxy-D-glucose, was also taken for comparative analysis against COVID-19 main protease protein (Figure 6).
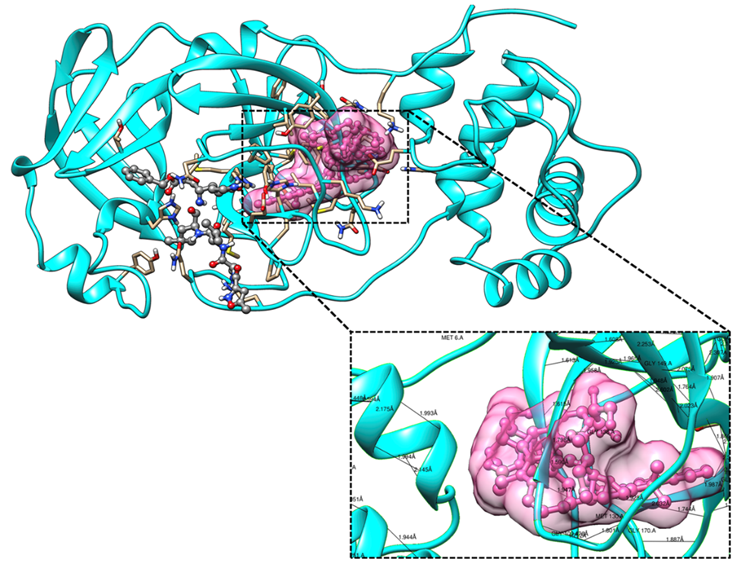
Figure 6: Molecular docking of the crystal structure of COVID-19 main protease complex (PDB ID:6LU7) with Cypate-2-Deoxy-D-glucose. The COVID-19 main protease complex was shown in cyan color in ribbon form, and modified 2-Desoxy-D-glucose was represented in a ball and stick model in the pink color surface. The enlarged version shows the binding mode of top-ranked docked poses and interacting critical residues of COVID-19 main protease with Cypate-2-Deoxy-D-glucose by forming hydrogen bonds. The docking was performed in AutoDock 4.2, and visualization of the complex was done by UCSF Chimera software.
The critical residues of the complex and the hydrophobic region were also generated in Ligplot software (Figure. 7). Moreover, the binding mechanism was more or less identical to the compounds mentioned above, with divergent orientations and positions of the compound. Although the affinity was found to be -9.18 kcal/mol, the binding energy with -8.01 kcal/mol indicated a less stable complex formation than the modified form. The exceptional residues such as Gln110, Thr111, Asn151 and Cys160 were found to be involved through hydrogen bonding with nitrogen and oxygen atoms. Potential Mpro inhibitors with high binding energy, such as saquinavir, aclarubicin, TMC-310911, and faldaprevir, contributed to the complex via hydrogen bonding by involving specific residues like Asn142, Ser144, and Gly143 [29]. By critically comparing the above three compounds on in silico basis screening, the modified 2-Deoxy-D-glucose was the best compound against the COVID-19 main protease complex. Recent studies discovered potential inhibitors against COVID-19 main protease, including the phytocompounds from Houttuynia cordata, marine bioactive compounds with antiviral activities, and marine polycyclic guanidine alkaloids [30,31]. Hence modified 2-Deoxy-D-glucose can be further examined as a suitable candidate drug against the critical condition of COVID-19. The results of this analysis may be fruitful in opening helpful research avenues against COVID during this pandemic.
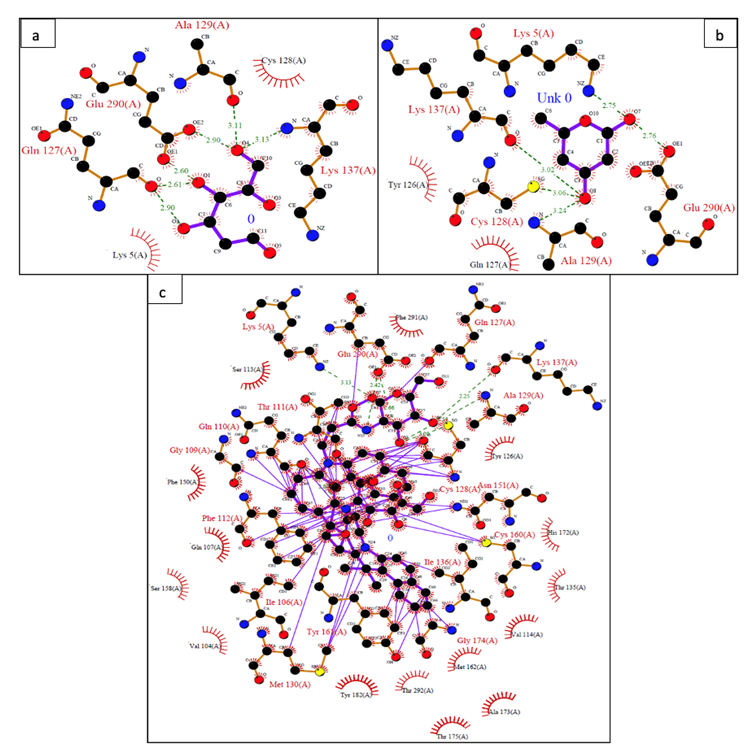
Figure 7: Interacting amino acid residues of COVID-19 main protease complex (PDB ID:6LU7) with (a) 2-Deoxy-D-glucose (b) modified 2-Deoxy-D-glucose and (c) Cypate-2-Deoxy-D-glucose was analyzed in LigPlot+ suite. The green lines indicate the hydrogen bonds and red dotted lines indicate the hydrophobic interactions. The bond formed between the atoms in purple color represents the ligand bonds and the bond in brown color represents the non-ligand bond.
ADMET prediction of 2-Deoxy-Glucose ligands for toxicity assessment
The growing demand for therapeutic strategy development and the discovery of competent drugs against COVID is essential in this pandemic. Consequently, the toxicity of these compounds was screened by the web server ADMETlab 2.0, and the properties were evaluated based on parameters like Absorption, Distribution, Metabolism, Excretion, and Toxicity. Regarding the physicochemical aspect, a few important parameters were considered vital in predicting efficient drug candidates against COVID-19 main protease. The detailed parameters are described in Table 3. The toxicity profile of 2-Deoxy-D-glucose and its various forms was evaluated based on a few potential parameters: molecular weight, which is critical for controlled and systematic drug delivery. These compounds' molecular weight was considered because they are crucial in the medication and oral bioavailability [32]. The analysis showed that the modified 2-Deoxy-D-glucose with the lowest molecular weight, i.e., 132.08 Da, may indicate rapid absorption and excretion rate compared to the other two compounds. Controlled and systematic delivery of low molecular weight drugs through brain barrier modulation prevented retinal degeneration and suppressed laser-induced choroidal neovascularisation in neurodegenerative conditions [33]. In addition, logP of the modified 2-Deoxy-D-glucose was within an optimal range from 0 to 3 while 2-Deoxy-D-glucose and Cypate 2-Deoxy-D-glucose were in the < 0 and > 3 range. According to the boundaries set by Lipinski’s Rule, no violation was found when the partition coefficient logP < 5 [34].
Table 3: Prediction of ADMET properties of 2-Deoxy-D-glucose, modified 2-Deoxy-D-glucose and cypate 2-Deoxy-D-glucose.
|
Parameters |
2-Deoxy-D-glucose |
Modified 2-Deoxy-D-glucose |
Cypate 2-Deoxy-D-glucose |
|
Molecular weight |
164.07 |
132.08 |
948.45 |
|
logP |
-2.035 |
0.029 |
3.383 |
|
logS |
-0.071 |
0.44 |
-3.999 |
|
H-Acceptors |
5 |
3 |
16 |
|
H-Donors |
4 |
2 |
10 |
|
Polar Surface Area |
97.99 |
49.69 |
244.98 |
|
HIA |
0.392 |
0.005 |
0.948 |
|
F30% |
0.911 |
0.513 |
0.981 |
|
Fu |
82.05% |
85.23% |
0.68% |
|
PPB |
22.54% |
6.58% |
97.58% |
|
CL |
1.954 |
8.07 |
1.13 |
|
T1/2 |
0.703 |
0.756 |
0.268 |
|
DILI |
0.02 |
0.047 |
0.552 |
|
Mutagenic |
none |
none |
4 |
The reduced bioavailability of the compound beyond these boundaries indicated poor absorption or permeation [35]. Similarly, the solubility of the compound, indicated by the logS value, improved the absorption at high rates. Apart from derivative and parent forms, the modified version evolved as an active inhibitor and increased the adsorption rate. The number of hydrogen bond acceptors and donors within the threshold values was ≤ 10 and ≤ 5, respectively. Another competent aspect, such as HIA, acted as an indicator of oral bioavailability, which was required to determine the efficacy of any drug [36]. Table 3 showed the efficacy of modified 2-Deoxy-D-glucose and concluded that the revised version was preferable to the other two. The HIA category is restricted from 0 to 1, whereas 0 to 0.3 resides in the excellent category, 0.3 to 0.7 belongs to the medium, and 0.7 to 1.0 belongs to the poor category [37]. Similar to HIA, F30% or human oral bioavailability by 30% was acted as a correlated factor and found to be within the moderate region, i.e., 0.513 for modified 2-Deoxy-D-glucose while the other two fell in the poor category (a range within 0-0.3: excellent; 0.3-0.7: medium; 0.7-1.0: poor). The fraction unbound in plasma, Fu, is critical for determining drug efficacy in drug pharmacokinetic studies because of its relation with unbound states, which alter the cellular response [38].
On the other hand, PPB may not significantly alter drug development strategy but must be considered when analyzing pharmacokinetic profiles in vivo conditions. The value of PPB ≤ 90% is considered a preferable category, while above 90% belongs to a poor level. The values were 6.58% of PPB in modified 2-Deoxy-D-glucose and 97.58% in Cypate. PPB has an influential impact on oral bioavailability/distribution and clearance of the drug. Earlier studies reported the consequence of high PPB on the antiviral activity of NVR 3-778, a sulfamoylbenzamide capsid assembly modulator [39]. The clearance of a drug for a modified one was found to be maximum. The threshold value of clearance of a drug is set by five, while more than it belongs to the favorable region and lesser to the low category [40]. Subsequently, half-life or T1/2 was related by clearance of the drug and was found to be within a moderate range for the molecules except for the derivative forms (0-0.3: excellent; 0.3-0.7: medium; 0.7-1.0: poor). The safety of these compounds was also evaluated, where both the parent and modified molecule had no mutagenic effect. However, the derivative molecule had a toxic effect. Similarly, the modified form's drug-induced liver injury or DILI effect was less than Cypate 2-Deoxy-D-glucose. A high level of DILI has serious clinical consequences that cause hepatotoxicity or acute liver failure and has been cited as one of the major reasons for drug removal from the market [41,42]. Thus, the modified 2-Deoxy-D-glucose had favorable ADMET parameters and could be considered a suitable drug candidate during the treatment of viral infection. However, using 2-Deoxy-D-glucose as an individual therapeutic agent associated with some risks, as long-term administration of high 2-Deoxy-D-glucose doses may cause toxicity in the individual. Furthermore, it has been demonstrated that a single therapeutic dose of the drug causes glucocytopenia in the nervous system, irregularities in the cardiovascular, respiratory, and immunological systems, and some normal tissues [43]. Thus, the study was conducted to enlighten the necessitates of targeting the virus during the early phase of infection is highly essential, and this can be achieved from rapid viral testing as a quick diagnosis to capture a therapeutic window of opportunity before an exuberant host-mediated immune response which leads to potentially fatal complications such as pneumonia, ARDS, multi-organ system dysfunction and hypercoagulation. Hence, by combining these parameters, the current findings indicated that the potentiality of 2-Deoxy-D-glucose was improved following structural modification. Thus, the modified forms have the potential to be used as a therapeutic strategy against virally infected cells by inhibiting or restricting energy production in viral cells. The in vivo study with the potential drug could be a game changer in the fight against pandemics like COVID. Early COVID-19 treatment can reduce disease recovery time, prevent hospitalizations, and be a more cost-effective way to manage COVID-19.
Conclusion
The current study investigated the potentiality of 2-Deoxy-D-glucose and its modified and derivative forms against the COVID-19 main protease complex. Compared to the derivative and parent compound, the report concluded that the modified form of 2-Deoxy-D-glucose by replacing the hydroxyl group with hydrogen atoms evolved as a promising candidate. The docking result demonstrated that the minimal energy of modified 2-Deoxy-D-glucose during the complex formation with COVID-19 main protease indicated the stability of the complex during the interaction. By supporting the docking results, ADMET analysis favored the potentiality of modified 2-Deoxy-D-glucose with minimal toxicity, while the derivative form of 2-Deoxy-D-glucose exhibited higher toxicity. Hence, the replacement of the hydroxyl group of 2-Deoxy-D-glucose can be used as a further effective strategy to treat the COVID-19 outbreak.
Declarations
Funding:
No funding was available
Conflicts of interest/Competing interests: All Authors express no conflicts of interest.
Authors Contributions
Conceptualisation, R.R.S., and A.K.D.; resources A.B.J, R.R.S., and S.S. writing—original draft preparation, R.R.S, and C.S.M.; writing—review and editing, R.R.S., A.K.D; supervision, A.K.D.; All authors have read and agreed to the published version of the manuscript.
Ethics approval (include appropriate permissions or waivers): Not applicable.
Consent to participate (include relevant statements): Not Applicable.
Consent for publication: Not applicable.
Availability of data and material (data transparency): Available on request.
Code availability (software application or custom code): Not applicable.
References
- Cimolai N. The Complexity of Co-Infections in the Era of COVID-19, SN Comprehensive Clinical Medicine 3 (2021): 1502-1514.
- Bandala ER, Kruger BR, Cesarino I, Leao AL, Wijesiri B, Goonetilleke A. Impacts of COVID-19 pandemic on the wastewater pathway into surface water: A review, Science of the Total Environment 774 (2021) 145586.
- Mi Q, Ma Y, Gao X, Liu R, Liu P, Mi Y, Fu X, Gao Q. 2-Deoxyglucose conjugated platinum (II) complexes for targeted therapy: design, synthesis, and antitumor activity 34 (2015) 2339-2350.
- Aygün ?, Kaya M, Alhajj R. Identifying side effects of commonly used drugs in the treatment of Covid 19, Scientific Reports 10 (2020): 1-14.
- Barlow A, Landolf KM, Barlow B, Yeung SYA, Heavner JJ, Claassen CW, et al. Review of Emerging Pharmacotherapy for the Treatment of Coronavirus Disease 2019, Pharmacotherapy 40 (2020): 416-437.
- Rathinavel T, Aroulmoji V. Hyaluronic Acid - 2-Deoxy-D-Glucose Conjugate Act as a Promising Targeted Drug Delivery Option for the Treatment of COVID-19, International Journal of Advanced Science and Engineering 7 (2021): 2-13.
- Mi Q, Ma Y, Gao X, Liu R, Liu P, Mi Y, et al. 2-Deoxyglucose conjugated platinum (II) complexes for targeted therapy: design, synthesis, and antitumor activity, Journal of Biomolecular Structure and Dynamics 34 (2016): 2339-2350.
- Verma A, Adhikary A, Woloschak G, Dwarakanath BS, Papineni RVL. A combinatorial approach of a polypharmacological adjuvant 2-deoxy-D-glucose with low dose radiation therapy to quell the cytokine storm in COVID-19 management, International Journal of Radiation Biology 96 (2020): 1323-1328.
- Dwarakanath BS. Cytotoxicity, radiosensitization, and chemosensitization of tumor cells by 2-deoxy-D-glucose in vitro., Journal of Cancer Research and Therapeutics 5 (2009): 27-31.
- Passalacqua KD, Lu J, Goodfellow I, Kolawole AO, Arche JR, Maddox RJ, et al. Glycolysis is an intrinsic factor for optimal replication of a norovirus, MBio 10 (2019).
- Fokt I, Ziemniak M, Ja A. 2-Deoxy-d-Glucose and Its Analogs: From Diagnostic to Therapeutic Agents (2020).
- Laussel C, Léon S. Cellular toxicity of the metabolic inhibitor 2-deoxyglucose and associated resistance mechanisms, Biochemical Pharmacology 182 (2020).
- Guo J, Du C, Shan L, Zhu H, Xue B, Qian Z, et al. Comparison of near-infrared fluorescent deoxyglucose probes with different dyes for tumor diagnosis in vivo, Contrast Media and Molecular Imaging 7 (2012): 289-301.
- Ye Y, Bloch S, Kao J, Achilefu S. Multivalent carbocyanine molecular probes: Synthesis and applications, Bioconjugate Chemistry 16 (2005): 51-61.
- Jin Z, Du X, Xu Y, Deng Y, Liu M, Zhao Y, et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors, Nature 582 (2020): 289-293.
- Santos KB, Guedes IA, Karl ALM, Dardenne LE. Highly Flexible Ligand Docking: Benchmarking of the DockThor Program on the LEADS-PEP Protein-Peptide Data Set, Journal of Chemical Information and Modeling 60 (2020): 667-683.
- Yang H, Yang M, Ding Y, Liu Y, Lou Z, Zhou Z, et al. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor, Proceedings of the National Academy of Sciences of the United States of America 100 (2003): 13190-13195.
- Anand K, Palm GJ, Mesters JR, Siddell SG, Ziebuhr J, Hilgenfeld R. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra α-helical domain, EMBO Journal 21 (2002): 3213-3224.
- Colovos C, Yeates TO. Verification of protein structures: Patterns of nonbonded atomic interactions, Protein Science 2 (1993): 1511-1519.
- Messaoudi A, Belguith H, Ben Hamida J. Homology modeling and virtual screening approaches to identify potent inhibitors of VEB-1 β-lactamase, Theoretical Biology and Medical Modelling 10 (2013): 1-10.
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures, Journal of Applied Crystallography 26 (1993): 283-291.
- Sisakht M, Mahmoodzadeh A, Darabian M. Plant-derived chemicals as potential inhibitors of SARS-CoV-2 main protease (6LU7), a virtual screening study, Phytotherapy Research (2021): 1-13.
- Hatada R, Okuwaki K, Mochizuki Y, Handa Y, Fukuzawa K, Komeiji Y, et al. Fragment Molecular Orbital Based Interaction Analyses on COVID-19 Main Protease - Inhibitor N3 Complex (PDB ID: 6LU7), Journal of Chemical Information and Modeling 60 (2020): 3593-3602.
- Stank A, Kokh DB, Fuller JC, Wade RC. Protein Binding Pocket Dynamics, Accounts of Chemical Research 49 (2016): 809-815.
- Guedes IA, Barreto AMS, Marinho D, Krempser E, Kuenemann MA, Sperandio O, et al. New machine learning and physics-based scoring functions for drug discovery, Scientific Reports 11 (2021): 1-19.
- Su JG, Jin Xu X, Hua Li C, Chen WZ, Wang CX. Identification of key residues for protein conformational transition using elastic network model, Journal of Chemical Physics 135 (2011).
- Choudhary MI, Shaikh M. Atia-Tul-Wahab, Atta-Ur-Rahman, In silico identification of potential inhibitors of key SARS-CoV-2 3CL hydrolase (Mpro) via molecular docking, MMGBSA predictive binding energy calculations, and molecular dynamics simulation, PLoS ONE 15 (2020): 1-15.
- Singh E, Khan RJ, Jha RK, Amera GM, Jain M, Singh RP, et al. A comprehensive review on promising anti-viral therapeutic candidates identified against main protease from SARS-CoV-2 through various computational methods, Journal of Genetic Engineering and Biotechnology 18 (2020).
- Keretsu S, Bhujbal SP, Cho SJ. Rational approach toward COVID-19 main protease inhibitors via molecular docking, molecular dynamics simulation and free energy calculation, Scientific Reports 10 (2020): 1-14.
- Das SK, Mahanta S, Tanti B, Tag H, Hui PK. Identification of phytocompounds from Houttuynia cordata Thunb. as potential inhibitors for SARS-CoV-2 replication proteins through GC-MS/LC-MS characterization, molecular docking and molecular dynamics simulation, Molecular Diversity (2021).
- El-Demerdash A, Metwaly AM, Hassan A, El-Aziz TMA, Elkaeed EB, Eissa IH, et al. Comprehensive virtual screening of the antiviral potentialities of marine polycyclic guanidine alkaloids against sars-cov-2 (Covid-19), Biomolecules 11 (2021): 1-25.
- Ya’u Ibrahim Z, Uzairu A, Shallangwa G, Abechi S. Molecular docking studies, drug-likeness and in-silico ADMET prediction of some novel β-Amino alcohol grafted 1,4,5-trisubstituted 1,2,3-triazoles derivatives as elevators of p53 protein levels, Scientific African 10 (2020): e00570.
- Campbell M, Humphries MM, Kiang AS, Nguyen ATH, Gobbo OL, Tam LCS, et al. Systemic low-molecular weight drug delivery to pre-selected neuronal regions, EMBO Molecular Medicine 3 (2011): 235-245.
- Schneider G. Prediction of Drug-Like Properties The “Drug-Likeness” Concept (2000): 1-34.
- Veber DF, Johnson SR, Cheng HY, Smith BR, Ward KW, Kopple KD. Molecular properties that influence the oral bioavailability of drug candidates, Journal of Medicinal Chemistry 45 (2002): 2615-2623.
- Yan A, Wang Z, Cai Z. Prediction of human intestinal absorption by GA feature selection and support vector machine regression, International Journal of Molecular Sciences 9 (2008): 1961-1976.
- Dong J, Wang NN, Yao ZJ, Zhang L, Cheng Y, et al. Admetlab: A platform for systematic ADMET evaluation based on a comprehensively collected ADMET database, Journal of Cheminformatics 10 (2018): 1-11.
- Watanabe R, Esaki T, Kawashima H, Natsume-Kitatani Y, Nagao C, Ohashi R, et al. Predicting Fraction Unbound in Human Plasma from Chemical Structure: Improved Accuracy in the Low Value Ranges, Molecular Pharmaceutics 15 (2018): 5302-5311.
- Helsen N, Vervoort T, Vandenbossche J, Lenz O, Monshouwer M, Pauwels F, et al. Effect of plasma protein binding on the anti-hepatitis B virus activity and pharmacokinetic properties of NVR 3-778, Antimicrobial Agents and Chemotherapy 62 (2018): 1-15.
- Xiong G, Wu Z, Yi J, Fu L, Yang Z, Hsieh C, et al. ADMETlab 2.0: an integrated online platform for accurate and comprehensive predictions of ADMET properties, Nucleic Acids Research (2021): 1-10.
- Walker PA, Ryder S, Lavado A, Dilworth C, Riley RJ. The evolution of strategies to minimise the risk of human drug-induced liver injury (DILI) in drug discovery and development, Archives of Toxicology 94 (2020): 2559-2585.
- Chen M, Suzuki A, Borlak J, Andrade RJ, Lucena MI. Drug-induced liver injury: Interactions between drug properties and host factors, Journal of Hepatology 63 (2015): 503-514.


 Impact Factor: * 6.2
Impact Factor: * 6.2 Acceptance Rate: 76.33%
Acceptance Rate: 76.33%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 