First Detection of NDM-type Carbapenemase-Producing K. pneumoniae isolated from Clinical Samples in Ouagadougou, Burkina Faso
Abdoul Karim Ouattara*, 1, 2, #, Blandine Ouédraogo1, #, Amana Mètuor Dabiré1, 3, 4, Rahimatou Yasmine Wendkuni Tiemtoré1, Serge Sougué1, Jacques Simporé1, 3
1Laboratoire de Biologie Moléculaire et de Génétique (LABIOGENE), UFR-SVT, Université Joseph KI-ZERBO, 03 BP 7021 Ouagadougou 03, Burkina Faso
2Université Norbert Zongo, Centre Universitaire de Manga, B.P. 376 Koudougou, Burkina Faso
3Centre de Recherche Biomoléculaire Pietro Annigoni (CERBA), 01 BP 364 Ouagadougou 01, Burkina Faso 4Université de Dédougou, BP 176 Dédougou, Dédougou, Burkina Faso
#These authors contributed equally to this work
*Corresponding author: Abdoul Karim Ouattara, Senior Lecturer in Genetics and Molecular Biology Teacher-Researcher, Université Norbert Zongo (UNZ), Centre Universitaire de Manga, B.P. 376 Koudougou, Burkina Faso
Received: 04 September 2023 Accepted: 14 September 2023 Published: 17 October 2023
Article Information
Citation: Abdoul Karim Ouattara, Blandine Ouédraogo, Amana Mètuor Dabiré, Rahimatou Yasmine Wendkuni Tiemtoré, Serge Sougué, Jacques Simporé. First Detection of NDM-type Carbapenemase-Producing K. pneumoniae isolated from Clinical Samples in Ouagadougou, Burkina Faso. Archives of Microbiology and Immunology. 7 (2023): 236-241
View / Download Pdf Share at FacebookAbstract
Background: Klebsiella pneumoniae is one of the most concerning Gram-negative opportunistic pathogens responsible for various infectious diseases. The rapid emergence and spread of clinically multidrug-resistant and hypervirulent strains of K. pneumoniae constitute a serious public health threat. The aim of this study was to identify the NDM-type carbapenemase gene in K. pneumoniae strains isolated from the Centre Hospitalier Universitaire Pédiatrique Charles De Gaulle (CHUP-CDG) de Ouagadougou, Burkina Faso.
Methods: Klebsiella pneumoniae strains were isolated from various biological samples (urine, pus, blood, stool, cerebrospinal fluid) between 2009 and 2013 at the CHUP-CDG. The antibiotic susceptibility test for ceftriaxone (CRO), ceftazidime (CAZ), cefotaxime (CTX), and imipenem (IMP) was performed using the disk diffusion method on Mueller-Hinton agar. The blaNDM gene was detected by classical PCR.
Results: Out of 10 Klebsiella pneumoniae strains, the susceptibility test showed high resistance to cephalosporins. The resistance rates were 80.00% for CRO, 90.00% for CTX, 100% for CAZ, and 10.00% for IMP. Molecular characterization of NDM-type metallo-β-lactamases by PCR revealed one (10.00%) strain carrying the blaNDM gene. Resistant strains and the NDM gene were mostly found in bacteria isolated from urine.
Conclusions: This study highlights the presence of the blaNDM gene in resistant strains of Klebsiella pneumoniae to β-lactams at CHUP-CDG. Surveillance measures should be taken to prevent the emergence of bacteria producing these enzymes.
Keywords
<p>Klebsiella pneumoniae, Antibiotic resistance, blaIMP gene, beta-lactamases, Burkina Faso</p>
Article Details
1. Introduction
The emergence of carbapenem resistance in Gram-negative bacilli represents a significant challenge as it leads to therapeutic dead-ends [1]. This resistance can be due to combined mechanisms involving β-lactamases with very low carbapenemase activity and decreased outer membrane permeability, or to the production of carbapenemases [2]. Carbapenemases are enzymes capable of hydrolyzing carbapenems [3]. Genes encoding these enzymes are often located on mobile genetic elements such as plasmids, transposons, integrons [4, 5] and can be transferred from one bacterial genus or species to another [6]. The most frequent carbapenemases are KPC-type enzymes, metallo-β-lactamases (VIM, IMP, NDM), and oxacillinases (OXA-48 and OXA-23) [7]. MBLs are characterized by the widest hydrolysis spectrum, able to hydrolyze all β-lactams except aztreonam [8]. The metallo-β-lactamase NDM-1 (New Delhi metallo-β-lactamase) was first identified in Sweden in a patient of Indian origin in early 2008 [3, 9]. Since then, it has received particular attention due to its rapid spread in Enterobacteriaceae [10]. Carbapenemase genes have been described in several epidemiological studies in Europe, North America, and Asia [11, 12]. In Africa, there is limited data available on carbapenemase genes. Previous studies have reported the presence of carbapenemase-producing bacteria in Southern, North, and West Africa [13]. n Burkina Faso, a study on bacteria resistance genes revealed the presence of the VIM- and IMP-2-encoded metallo-β-lactamase [14]. Our research team also identified several bacterial strains producing extended-spectrum β-lactamases with multidrug resistance in Burkina Faso [15-18], Togo [19-21] and Niger [22]. However, data on NDM-type metallo-β-lactamase in resistant bacteria is scarce. The main objective of this study was to characterize the NDM-type carbapenemase gene carried by Klebsiella pneumoniae strains isolated between 2009-2013 at the Centre Hospitalier Universitaire Pédiatrique Charles De Gaulle (CHUP-CDG) de Ouagadougou, Burkina Faso.
2. Materials and Methods
2.1 Type and period of study
This was a descriptive study with retrospective collection of bacterial strains [15, 23]. The study was conducted in Ouagadougou, at the Centre de Recherche Biomoléculaire Pietro Annigoni (CERBA) and the Laboratoire de Biologie Moléculaire et de Génétique (LABIOGENE) of Université Joseph Ki-Zerbo, between June and September 2021.
2.2 Sampling
A total of 10 strains of K. pneumoniae responsible for human infection were included in this study. These strains were isolated between 2009 and 2013 from various biological samples such as urine, pus, and cerebrospinal fluid at the CHUP-CDG de Ouagadougou, Burkina Faso.
2.3 Antimicrobial susceptibility testing
The antibiotic resistance of the strains was tested using the disk diffusion method on Mueller-Hinton agar (MH) in accordance with the recommendations of the Antibiogram Committee of the French Microbiology Society [24]. The antibiotics ceftriaxone (CRO), ceftazidime (CAZ), cefotaxime (CTX), and imipenem (IMP) were used.
2.4 DNA extraction
DNA extraction was performed by the boiling method [16]. An isolated colony was picked from the MH agar plates and suspended in 200 µl of distilled water in labeled Eppendorf tubes. The bacterial suspension was then placed in a water bath at 100 °C for 15 minutes to break the bacterial wall and membrane and release the genetic material. The tubes were finally centrifuged at 12,000 rpm for 10 minutes, and the supernatant containing the total DNA was transferred to a new Eppendorf tube and stored at -20 °C until molecular analyses.
2.5 Detection of the blaNDM gene by PCR
The blaNDM gene was detected by PCR using the GeneAmp® PCR System 9700 thermocycler (Applied Biosystems, California, USA) in a reaction volume of 20 μL. The reaction mixture consisted of 4 μL of GREEN PCR Master Mix, 0.5 μL of forward and reverse primers, 14 μL of PCR water, and 1 μL of bacterial DNA from each strain. The amplification of the blaNDM gene was carried out using the following primer pair: F-5’CCATGCGGGCCGTATGAGTGATT’3 and R-5’AAGCTGAGCACCGCATTAGCCG’3 with an expected amplicon of 763 bp. A negative control was prepared using 19 μL of reaction mix and 1 μL of PCR water. The following PCR program was used: an initial denaturation step at 96 °C for 5 minutes, followed by 30 cycles, each consisting of denaturation at 96 °C for 30 seconds, annealing at 62 °C for 30 seconds, and extension at 72 °C for 30 seconds. A final extension step was performed at 72 °C for 7 minutes.
2.6 Agarose gel electrophoresis
The DNA fragments amplified by PCR were separated by electrophoresis on a 1.5% agarose gel prepared in a 1X tris-borate-EDTA solution and containing 0.5 μg/mL of ethidium bromide. Migration was performed at 100 V for 25 minutes and the amplicons were visualized under UV light using the GeneFlash device (Syngene, Bio-Imaging, UK).
2.7 Ethical considerations
The institutional ethics committee of CERBA/LABIOGENE reviewed and approved the protocol of this study.
2.8 Statistical analysis
The collected data were entered into Excel 2016 and analyzed using the STATA software. The different results were expressed as percentages (%) and frequency for categorical variables.
3. Results
3.1 Distribution of bacterial strains by pathological samples
The 10 Klebsiella pneumoniae strains analyzed in this study were obtained from children at CHUP-CDG and were isolated from various pathological samples. Among the 10 strains, the majority (50%, 5/10) were isolated from urine samples, followed by pus samples (40%, 4/10), and cerebrospinal fluid (10%, 1/10).
3.2 Antibiotic resistance and detection of the blaNDM gene
Table I presents the distribution of resistance among biological samples, as determined by the sensitivity test of the 10 clinical strains of Klebsiella pneumoniae in the present study. The results indicate a high level of resistance to cephalosporins, with rates of 80.00% (8/10) for ceftriaxone (CRO), 90.00% (9/10) for cefotaxime (CTX), and 100% (10/10) for ceftazidime (CAZ). In contrast, only 10.00% (1/10) of the strains exhibited resistance to imipenem (IMP).
Table I: Distribution of resistance by biological sample
|
Species |
CTX |
CRO |
CAZ |
IMP |
|
Urines |
5 (50.00 %) |
4 (40.00 %) |
5 (50.00 %) |
1 (10.00 %) |
|
Pus |
3 (30.00 %) |
3 (30.00 %) |
4 (40.00 %) |
0 (00 %) |
|
CSF |
1 (10.00 %) |
1 (10.00 %) |
1 (10.00 %) |
0 (00 %) |
|
Total |
9 (90.00 %) |
8 (80.00 %) |
10 (100.00 %) |
1 (10.00 %) |
CSF: Cerebrospinal fluid, CTX: Cefotaxime; CAZ: Ceftazidime; CRO: Ceftriaxone, IMP: Imipenem
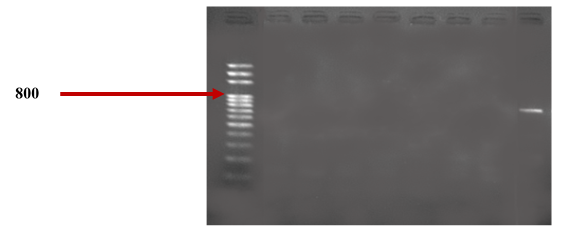
Molecular characterization of NDM-type metallo-β-lactamases by PCR revealed that 10.00% (1/10) of the strains carried the blaNDM gene. The urine samples yielded the strain carrying the blaNDM gene, as shown in table 2. The electrophoretic profile of the blaNDM gene is illustrated in figure 1.
Table 2: Presence of blaNDM gene according to the biological sample
|
Biological samples |
blaNDM gene |
|
Urines |
1 (10. 00 %) |
|
Pus |
0 (00 %) |
|
CSF |
0 (00 %) |
|
Total |
1 (10. 00 %) |
CSF: Cerebrospinal fluid, CTX: Cefotaxime; CAZ: Ceftazidime; CRO: Ceftriaxone, IMP: Imipenem
4. Discussion
Carbapenem resistance in Enterobacteriaceae due to carbapenemase production is a major public health problem [2]. Infections caused by carbapenemase-producing bacteria lead to true therapeutic impasses [1]. Carbapenems are a subclass of β-lactams, which also includes cephalosporins, monobactams, and penicillins [25]. The aim of the present study was to determine the frequency of the blaNDM gene by PCR in clinical isolates of K. pneumoniae. Most of these clinical strains were isolated from urine and pus. Several previous studies conducted by our research team have also reported a predominance of multidrug-resistant strains in urine samples in Burkina Faso [16-18, 23], Togo [19-21] and Niger [22]. Urinary tract infection is the second most common bacterial infection in children and is considered a threat to public health due to the increasing rates of antibiotic resistance among uropathogens [26]. The antibiotic susceptibility test of the strains in this study revealed a resistance rate of 80.00% for ceftriaxone (CRO), 90.00% for cefotaxime (CTX), and 100% for ceftazidime (CAZ) among third generation cephalosporins. Cephalosporins are commonly prescribed in outpatient and inpatient settings, and their broad spectrum of activity allows for diverse use in most medical specialties [27]. Currently, there are five generations of cephalosporins, primarily differentiated by their structure, spectrum of activity, and side effect profiles [28]. Like other β-lactams, cephalosporins act by preventing bacteria from forming their cell wall, leading to their death [29]. Resistance to third-generation cephalosporins among Gram-negative bacilli has been described in several studies in Burkina Faso [14, 18, 30] and in the West African sub-region[31, 32]. β-lactams are among the most commonly prescribed antibiotics in the world, with the emergence of multidrug resistance through several mechanisms or the production of several types of enzymes capable of inactivating these drugs [33]. The present study found a lower resistance rate (10%, 1/10) to imipenem. Resistance to carbapenems has been reported in Asia with a 1.20% resistance rate to meropenem [34] and 65.6% for imipenem [35], in Europe with a 24.32% resistance rate to imipenem [36], in West Africa with 100% resistance rate to meropenem [37] and in Southern Africa with a 96.7% resistance rate to meropenem [38]. The production of carbapenemases, overexpression of efflux pumps, as well as mutations or repression of porins are all mechanisms of resistance against imipenem [39]. The search for the resistance gene showed a strain carrying blaNDM. NDM can hydrolyze a broad spectrum of β-lactams, including penicillin, cephalosporins, and carbapenems. The presence of the blaNDM gene in K. pneumoniae strains has been reported in several African countries such as Kenya [40], Tanzania [41] and Sudan [42]. The emergence of clinical strains of Klebsiella pneumoniae co-producing KPC-2 and NDM-1 and resistant to carbapenems has also been reported in the literature [43], suggesting ongoing surveillance efforts and the imperative need for new therapeutic solutions to combat the expansion of multidrug resistance.
5. Conclusion
The present study reports a clinical strain of K. pneumoniae that is multidrug-resistant and carries the blaNDM gene. Carbapenem resistance in Enterobacteriaceae is a major public health problem, particularly in developing countries such as Burkina Faso, where it is associated with therapeutic dead-ends. This suggests that adequate surveillance measures are needed to prevent the emergence and dissemination of this resistance gene in other clinical strains.
Declarations
Acknowledgments
The authors wish to thank all participants in this study. A deep gratitude to all the staff of Saint Camille Hospital of Ouagadougou (HOSCO) and Biomolecular Research Center Pietro Annigoni (CERBA) for technical support.
Funding
No funding was received for this study.
Authorship Contributions
Study concept and design: AKO, AMD, RYWT and JS. Study execution and acquisition of data: AKO, BO, AMD, RYWT and SS. Statistical analysis and interpretation of data: AKO, BO, AMD. Drafting of the manuscript: AKO, BO and AMD. Critical revision of the manuscript for important intellectual content: AKO, BO, AMD, RYWT, AMD, SS and JS. Administrative, technical, and material support: AKO, AMD, RYWT and SS. Study supervision: AKO, RYWT, AMD, and JS. The Corresponding Author declare that the manuscript has been read and approved by all named authors and that the order of authors listed in the manuscript has been approved by all of us.
Competing interests
The authors declare no conflict of interest.
References
- Jean, SS, Harnod, D, and Hsueh, PR. Global Threat of Carbapenem-Resistant Gram-Negative Bacteria. Front Cell Infect Microbiol (2022): 823684.
- Nordmann, P and Poirel, L. Epidemiology and Diagnostics of Carbapenem Resistance in Gram-negative Bacteria. Clinical Infectious Diseases (2019): S521-S528.
- Wu W, Feng Y, Tang G, Qiao F, McNally A, and Zong Z. NDM Metallo-β-Lactamases and Their Bacterial Producers in Health Care Settings. Clinical Microbiology Reviews (2019): e00115-18.
- Diene, SM and Rolain, JM. Carbapenemase genes and genetic platforms in Gram-negative bacilli: Enterobacteriaceae, Pseudomonas and Acinetobacter species. Clinical Microbiology and Infection (2014): 831-838.
- Yang TY, Lu PL, and Tseng SP. Update on fosfomycin-modified genes in Enterobacteriaceae. J Microbiol Immunol Infect (2019): 9-21.
- Salamzade R, Manson AL, Walker BJ, Brennan-Krohn T, Worby CJ, Ma P, et al. Inter-species geographic signatures for tracing horizontal gene transfer and long-term persistence of carbapenem resistance. Genome Medicine (2022): 37.
- Foudraine DE, Dekker LJM, Strepis N, Bexkens ML, Klaassen CHW, Luider TM, et al. Accurate Detection of the Four Most Prevalent Carbapenemases in E. coli and K. pneumoniae by High-Resolution Mass Spectrometry. Front Microbiol (2019): 2760.
- Tzouvelekis LS, Markogiannakis A, Psichogiou M, Tassios PT, and Daikos GL. Carbapenemases in Klebsiella pneumoniae and Other Enterobacteriaceae: an Evolving Crisis of Global Dimensions. Clinical Microbiology Reviews (2012): 682-707.
- Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, et al. Characterization of a New Metallo-β-Lactamase Gene, <i>bla</i> <sub>NDM-1</sub> , and a Novel Erythromycin Esterase Gene Carried on a Unique Genetic Structure in <i>Klebsiella pneumoniae</i> Sequence Type 14 from India. Antimicrobial Agents and Chemotherapy (2009): 5046-5054.
- Bush K and Bradford PA. Epidemiology of β-Lactamase-Producing Pathogens. Clin Microbiol Rev (2020).
- Yu H, Ezpeleta-Lobato G, Han X, Carmona-Cartaya Y, and Quiñones-Pérez D. Carbapenamase-Producing Acinetobacter baumannii in China, Latin America and the Caribbean. MEDICC Rev (2022): 59-69.
- Chen HY, Jean SS, Lee YL, Lu MC, Ko WC, Liu,PY, et al. Carbapenem-Resistant Enterobacterales in Long-Term Care Facilities: A Global and Narrative Review. Front Cell Infect Microbiol (2021): 601968.
- Ouedraogo AS, Jean Pierre H, Bañuls AL, Ouédraogo R, and Godreuil S. Emergence and spread of antibiotic resistance in West Africa: contributing factors and threat assessment. Med Sante Trop (2017): 147-154.
- Sanou S, Ouedraogo AS, Aberkane S, Vendrell J, Ouchar O, Bouzimbi N, et al. Prevalence and Molecular Characterization of Extended Spectrum β-Lactamase, Plasmid-Mediated Quinolone Resistance, and Carbapenemase-Producing Gram-Negative Bacilli in Burkina Faso. Microb Drug Resist (2021): 18-24.
- Metuor Dabire A, Zongo KJ, Zeba B, Moussawi J, Baucher M, and El Jaziri M. Resistances to the Oxyimino-Cephalosporins by Ctx-M-15 Producing Klebsiella Isolated From the Urines Samples of Patients in the University Hospital Complex Paediatric Charles De Gaulle (CHUP-CDG) Of Ouagadougou in Burkina Faso Journal of Asian Scientific Research (2013): 882-890.
- Mètuor DA, Tiemtoré RYW-K, Bangré YA, Zohoncon TM, Sougué S, Zongo JK, et al. Detection of multidrug-resistant enterobacteria simultaneously producing extended-spectrum -lactamases of the PER and GES types isolated at Saint Camille Hospital Center, Ouagadougou, Burkina Faso. African Journal of Microbiology Research (2019): 414-420.
- Tiemtoré RY, Mètuor-Dabiré A, Zohoncon TM, Bangré YA, Sougue S, Zongo J, et al. First Detection of PE-Type Extended-Spectrum β-lactamases at Saint Camille Hospital Center of Ouagadougou, Burkina Faso. Int. J. Biochem. Biophys. Mol. Biol (2019).
- Tiemtoré RYW, Mètuor Dabiré A, Ouermi D, Sougué S, Benao S, and Simporé, J. Isolation and Identification of Escherichia coli and Klebsiella pneumoniae Strains Resistant to the Oxyimino-Cephalosporins and the Monobactam by Production of GES Type Extended Spectrum Bêta-Lactamase (ESBL) at Saint Camille Hospital Center in Ouagadougou, Burkina Faso. Infect Drug Resist (2022): 3191-3204.
- Diagbouga S, Salah F, Sadji A, Dabire A, Nadembega, C, Kere, A, et al. Detection of High Prevalence of TEM/SHV/CTX-M Genes in ESBL Producing and Multidrug Resistant Klebsiella pneumoniae and Klebsiella oxytoca. J Clin Diagn Res (2016): 000129.
- Salah F, Diagbouga S, Dabire AM, Sadji A, Nadembega C, and Moumouni, A. First detection of Resistance genes encoding extended Spectrum β-lactamase producing Escherichia coli at Lomé, Togo. Archives of Clinical Microbiology (2016): 32.
- Salah FD, Soubeiga ST, Ouattara AK, Sadji AY, Metuor-Dabire A, Obiri-Yeboah D, et al. Distribution of quinolone resistance gene (qnr) in ESBL-producing Escherichia coli and Klebsiella spp. in Lomé, Togo. Antimicrob Resist Infect Control (2019): 104.
- Moumouni A, Diagbouga S, Nadembèga C, Metuor Dabire A, Soubeiga ST, Ouattara AK, et al. Quinolone Resistance (qnr) genes in fecal carriage of extended Spectrum beta-lactamases producing Enterobacteria isolated from children in Niger. Curr Res Microbiol Biotechnol (2017): 953-7.
- Metuor Dabire A, Zongo KJ, Zeba B, Traoré/Ouedraogo R, Moussawi J, Baucher M, et al. First detection of SHV-type extended spectrum β-lactamases in the University Hospital Complex Paediatric Charles De Gaulle (CHUP-CDG) of Ouagadougou in Burkina Faso. Journal of Asian Scientific Research (2014): Journal of Asian Scientific Research.
- CA-SFM/EUCAST, Comité de l’antibiogramme de la Société Française de Microbiologie - Recommandations 2021 V.01 Avril (2021).
- El-Gamal MI, Brahim I, Hisham N, Aladdin R, Mohammed H, and Bahaaeldin A. Recent updates of carbapenem antibiotics. Eur J Med Chem (2017): 185-195.
- Millner R and Becknell, B. Urinary Tract Infections. Pediatr Clin North Am (2019): 1-13.
- Jordan E, Voide C, Petignat PA, and Gobin, N. [Cephalosporins in clinical practice]. Rev Med Suisse (2020): 1906-1911.
- Fernandez J, Jimenez-Rodriguez TW, and Blanca-Lopez, N. Classifying cephalosporins: from generation to cross-reactivity. Curr Opin Allergy Clin Immunol (2021): 346-354.
- Lin X and Kück U. Cephalosporins as key lead generation beta-lactam antibiotics. Appl Microbiol Biotechnol (2022): 8007-8020.
- Kpoda DS, Ajayi A, Somda M, Traore O, Guessennd N, Ouattara AS, et al. Distribution of resistance genes encoding ESBLs in Enterobacteriaceae isolated from biological samples in health centers in Ouagadougou, Burkina Faso. BMC Research Notes (2018): 471.
- Toudji AG, Djeri B, Karou SD, Tigossou S, Ameyapoh Y, and De Souza C. Prévalence des souches d’entérobactéries productrices de bêta-lactamases à spectre élargi isolées au Togo et de leur sensibilité aux antibiotiques. International Journal of Biological and Chemical Sciences (2017): 1165.
- Mofolorunsho KC, Ocheni HO, Aminu RF, Omatola CA, and Olowonibi OO. Prevalence and antimicrobial susceptibility of extended-spectrum beta lactamases-producing Escherichia coli and Klebsiella pneumoniae isolated in selected hospitals of Anyigba, Nigeria. Afr Health Sci (2021): 505-512.
- Munita JM and Arias CA. Mechanisms of Antibiotic Resistance. Microbiol Spectr (2016).
- Nirwati H, Sinanjung K, Fahrunissa F, Wijaya F, Napitupulu S, Hati VP, et al. Biofilm formation and antibiotic resistance of Klebsiella pneumoniae isolated from clinical samples in a tertiary care hospital, Klaten, Indonesia. BMC Proceedings (2019): 20.
- Effah CY, Sun T, Liu S, and Wu Y. Klebsiella pneumoniae: an increasing threat to public health. Annals of Clinical Microbiology and Antimicrobials (2020): 1.
- Ballén V, Gabasa Y, Ratia C, Ortega R, Tejero M, and Soto S. Antibiotic Resistance and Virulence Profiles of Klebsiella pneumoniae Strains Isolated from Different Clinical Sources. Frontiers in Cellular and Infection Microbiology (2021): 738223.
- Obeng-Nkrumah N, Twum-Danso K, Krogfelt KA, and Newman MJ. High Levels of Extended-Spectrum Beta-Lactamases in a Major Teaching Hospital in Ghana: The Need for Regular Monitoring and Evaluation of Antibiotic Resistance. The American Journal of Tropical Medicine and Hygiene (2013): 960-964.
- Teklu DS, Negeri AA, Legese MH, Bedada TL, Woldemariam HK, and Tullu KD. Extended-spectrum beta-lactamase production and multi-drug resistance among Enterobacteriaceae isolated in Addis Ababa, Ethiopia. Antimicrobial Resistance & Infection Control (2019): 39.
- Li S, Wu J, Ma N, Liu W, Shao M, Ying N, et al. Prediction of genome-wide imipenem resistance features in Klebsiella pneumoniae using machine learning. Journal of Medical Microbiology (2023).
- Poirel L, Revathi G, Bernabeu S, and Nordmann P. Detection of NDM-1-Producing <i>Klebsiella pneumoniae</i> in Kenya. Antimicrobial Agents and Chemotherapy (2011): 934-936.
- Mushi MF, Mshana SE, Imirzalioglu C, and Bwanga F. Carbapenemase Genes among Multidrug Resistant Gram Negative Clinical Isolates from a Tertiary Hospital in Mwanza, Tanzania. BioMed Research International (2014): 1-6.
- Adam MA and Elhag WI. Prevalence of metallo-β-lactamase acquired genes among carbapenems susceptible and resistant Gram-negative clinical isolates using multiplex PCR, Khartoum hospitals, Khartoum Sudan. BMC Infectious Diseases (2018): 668.
- Gao H, Liu Y, Wang R, Wang Q, Jin L, and Wang H. The transferability and evolution of NDM-1 and KPC-2 co-producing Klebsiella pneumoniae from clinical settings. EBioMedicine (2020): 102599.


 Impact Factor: * 3.5
Impact Factor: * 3.5 Acceptance Rate: 71.36%
Acceptance Rate: 71.36%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 
