In Vitro and In Vivo Evaluation of Antibiotic Combinations against Multidrug Resistant Proteus Mirabilis Isolated from Admitted Patients of Dhaka Medical College Hospital, Dhaka
Rizwana Zaman1 *, S.M. Shamsuzzaman2
1Lecturer, department of microbiology, Dhaka medical college, Dhaka, Bangladesh
2Head, department of microbiology, Dhaka medical college, Dhaka, Bangladesh
*Corresponding author: Rizwana Zaman, Lecturer, Department of microbiology, Dhaka medical college, Dhaka, Bangladesh
Received: 12 July 2021; Accepted: 20 July 2021; Published: 03 August 2021
Article Information
Citation: Rizwana Zaman, S.M. Shamsuzzaman. In vitro and in vivo evaluation of antibiotic combinations against multidrug resistant Proteus mirabilis isolated from admitted patients of Dhaka Medical College Hospital, Dhaka. Archives of Microbiology and Immunology 5 (2021): 243-262.
View / Download Pdf Share at FacebookAbstract
Emergence of MDR P. mirabilis is a threat for high morbidity and mortality with catheter-associated urinary tract infections leading to urolithiasis and permanent renal damage, bacteremia and sepsis. Global shortage of new effective antibiotic with reduced susceptibility of P. mirabilis to imipenem, tigecycline and resistance to colistin needs antimicrobial combinations. So, different antimicrobial combinations were used to see their efficacy both in vitro and in vivo with their resistance in Bangladesh.
MIC of antibiotics alone and in combination and efficacies of different antibiotic combinations in vivo (mice model) were evaluated. PCR and sequencing of resistance genes were done.
Among the 500 samples, 70% were culture positive and out of these, 10.57% were P. mirabilis. Among isolated P. mirabilis, 24.32%, 21.62%, 27.03% and 78.38% were resistant to fosfomycin, imipenem, tigecycline and amikacin respectively and 75.68%, 13.51% and 10.81% were MDR, XDR and PDR respectively. Among fosfomycin resistant P. mirabilis, 77.78%, 44.44% and 22.22% were positive for fosA, fosA3 and fosA4 respectively. Among imipenem resistant P. mirabilis, 62.50% were positive for NDM-1 and OXA-10, 50% for NDM-2, OXA-23 and OXA-48, 25% for OXA-58.
In vitro combination of imipenem-amikacin and imipenem-fosfomycin showed 100%, and 75% synergism respectively. The best in vivo results appeared in the group treated with imipenem-amikacin and imipenem-fosfomycin combinations where 100% bacterial clearance was observed.
As P. mirabilis showed high level resistance towards commonly used antimicrobial agents, it is very difficult to treat by a single agent. So, emphasis should be given on using antibiotic combinations.
Keywords
<p>Antibiotic combination, FIC, MDR, MIC, PDR, XDR</p>
Article Details
Highlights:
Drug combinations are more efficacious than single drug therapy
1. Introduction
The emergence and spread of carbapenem resistant Enterobacteriaceae have become an increasing concern for healthcare services worldwide. Infections caused by these bacteria including P. mirabilis cause significant morbidity and mortality [3]. Broadly carbapenemases are categorized into three groups: Class A (penicillinases), Class B (metallo-β-lactamases [MBL]), and Class D (Oxacillinases) [6]. The New Delhi metallo-β-lactamase (NDM) is one of the class B metallo-β-lactamases. They are present largely inEnterobacteriaceae [30]. Decreased susceptibility to imipenem, intrinsic resistance to polymyxins and tetracyclines along with the acquisition of β-lactam resistance traits by P. mirabilis seriously limits treatment options [34].
Recently, the use of fosfomycin has attracted renewed interest for the treatment of serious systemic infections caused by multidrug-resistant Enterobacteriaceae [38]. The WHO has classified fosfomycin in the category of a ‘critically important’ and has been identified as an antimicrobial which holds great promise worldwide for MDR gram negative infection, due to its affordable cost and efficacy as carbapenem sparing regimen [45]. It is among the limited treatment options for carbapenem resistant Enterobacteriaceae (CRE) infections [2].
Although the contribution of fosfomycin inactivating enzymes in emergence and spread of fosfomycin resistance currently seems low-to-moderate, their presence in transferable plasmids may potentially provide the best means for the spread of fosfomycin resistance in the future [46, 29].
So, until better antibiotics are being developed, novel antibiotic combinations yield some in vitro and in vivo synergistic activity are perhaps the best options we have in our hands to manage multidrug resistance. Combination of two antibiotics may provide broader spectrum coverage, decrease the emergence of resistance & also dose related toxicity [24]. In addition, animal models provide not only an important resource for the development and testing of new therapies, but also for studies into the initiation and maintenance of bacterial infections [22].
To the best of knowledge, no study has been carried out in Bangladesh regarding use of different antibiotic combinations on multidrug resistant Proteus mirabilis both in vitro and in vivo. This study seeks to determine multidrug resistant Proteus mirabilis among patients of DMCH and to evaluate the effectiveness of different antibiotic combinations both in vitro and in vivo (in mouse model) along with their PCR assays.
2. Methods
2.1 Clinical isolates and identification
A cross sectional study was conducted from January 2019 to December 2019 among 500 samples of urine, wound swab, pus and blood of adult patients having clinically suspected infections admitting in Dhaka Medical College Hospital or were received in the microbiology department for culture and sensitivity after taking informed written consent irrespective of sex and antibiotic intake. Patients who did not give consent were excluded from this study. Out of these 500 samples, 350 (70%) samples were culture positive, of which 37 (10.57%) were identified as Proteus mirabilis by cultural, biochemical and phenotypic tests.
2.2 Antimicrobial susceptibility test
Susceptibility to antimicrobial agents of all isolated organisms were determined by Kirby-Baurer modified disc diffusion technique using Mueller-Hinton plates and zones of inhibition were interpreted according to CLSI guidelines [9]. The criteria for the United States Food and Drug Administration was used for the interpretation of zone of inhibition of tigecycline. Antibiotic discs were obtained from commercial sources (Oxoid Ltd, UK). Following antimicrobial discs were used: amikacin (30μg), piperacillin-tazobactam (100/10μg), imipenem (10μg), tigecycline (15µg), ciprofloxacin (30μg), cefepime (30μg), ceftazidime (30μg), ceftriaxone (30μg), cefoxitin (30μg), cefuroxime sodium (30μg), amoxiclav (amoxicillin 20μg & clavulanic acid 10μg) and aztreonam (10μg). Fosfomycin susceptibility was tested by agar dilution method of MIC.
2.3 Agar dilution method of MIC
MIC of imipenem, piperacillin-tazobactam (Reneta Pharma Limited, Dhaka), tigecycline (Incepta Pharma Limited, Dhaka), amikacin (ACI Pharma Limited, Dhaka), fosfomycin (Beximco Pharma Limited) was determined by agar dilution method [1].
2.3.1. Inoculums preparation & Inference of MIC
As 0.5 McFarland turbidity standard contains 1×108 cfu/ml [7]. 10 times dilution of test inoculums was done to achieve 1× 107 cfu/ml. All the inoculated plates were incubated aerobically at 37ºC overnight. The lowest concentration of antibiotic impregnated Mueller-Hinton agar showing no visible growth on agar media was considered as MIC of the drug of that strain of bacteria. Escherichia coli ATCC 25922 were used as control organisms.
2.3.2. Effects of antibiotic combinations in vitro
Combination of antibiotics was done to see synergistic, additive, indifference or antagonistic effects by agar dilution method. By the agar dilution method synergy was present when there was a fourfold or greater reduction in the MICs of both antibiotics. A reduction of less than fourfold in the MICs of both antibiotics were considered additive. Indifference was found when neither drug exhibited a decrease in MIC and an increase in the MIC was considered antagonism [18].
Synergy between different antibiotic combinations also was assessed by fractional inhibitory concentration (FIC) by using the formula of FICI (Fractional inhibitory concentration index) by agar dilution method. So, FICI =
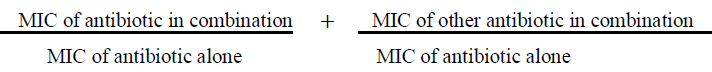
The FICI were interpreted as follows: Synergy defined when FICI value ≤ 0.5; additivity, FICI of >0.5 to ≤1; indifference, FICI of >1 to ≤4; antagonism, FICI of >4 [39].
2.3.3. Preparation of Mueller Hinton agar plate containing combination of two antibiotics
For each sample, four plates were prepared with 50ml Mueller Hinton agar media in each plate. The first plate contained the MIC value of two antibiotics, for that sample. The second plate contained two folds lower dilution than the MIC of two antibiotics, for that sample. The third plate contained four folds lower dilution than the MIC of both antibiotics and the fourth plate contained eight folds lower dilution than the MIC of both antibiotics for that sample. The Mueller-Hinton agar plate was impregnated with antibiotic stock solution according to the above description.
2.4. Molecular method [13, 21]
Polymerase chain reaction (PCR) was done for the detection of multidrug resistance genes in Proteus mirabilis.
2.4.1. Procedure of Bacterial pellet formation
A loop full of bacterial colonies from MHA media was inoculated into a microcentrifuge tube having sterile TSB and incubated overnight at 37ºC. Incubated tube was centrifuged at 4000g for 10 minutes. Supernatant was discarded and tubes containing bacterial pellets were kept at -20ºC for DNA extraction.
2.4.2. DNA extraction
Three hundred microlitre of sterile distilled water was added to microcentrifuge tubes having pellets and vortexed until mixed well. Then the mixture was heated at 100° C for 10 minutes in a heat block. After heating, tubes were immediately placed on ice for 5 minutes and centrifuged at 14000 g for 6 minutes at 4ºC. Finally, the supernatant was taken into another microcentrifuge tube. This extracted DNA was preserved at 4ºC for 7-10 days and -20ºC for a long time.
2.4.3. Mixing of mastermix with primer and DNA template
PCR was performed in a final reaction volume 25 µl in a PCR tube, containing 12.5 µl of master mix (mixture of dNTP, taq polymerase, MgCl2 and PCR buffer), 2 µl forward primer, 2 µl reverse primer (Promega Corporation, USA) , 2 µl of extracted DNA and 6.5 µl of nuclease free water. After a brief vortex, the tubes were centrifuged.
2.4.4. Amplification in thermal cycler (Gene Atlas, Master cycler gradient, Japan, Model482)
PCR assays were performed in a DNA thermal cycler. After amplification products were processed for gel documentation or kept at -20ºC till tested.
2.4.5. Agarose gel electrophoresis and visualization
PCR products were detected by electrophoresis on 1.5% agarose gel. Gel was prepared with 1 X TBE buffer (Tris EDTA). For 1.5% agarose gel preparation, 0.18 gram agarose powder (LE, analytic grade, Promega, Madison, USA) was mixed with a 1.25 ml TBE buffer. A comb was placed in a gel tray, the gel was poured. After solidification, 1 µl of loading dye and 5 µl of amplicon was mixed on parafilm and was loaded in agarose well. Similarly, 2 µl of 100bp DNA ladder was mixed with 1µl loading dye and was loaded. Gel electrophoresis was done in 230 voltages for 30 minutes. After electrophoresis, the gel was stained with ethidium bromide (20µl ethidium bromide in 200 ml distilled water). The gel was observed under UV transilluminator (Gel Doc, Major Science, Taiwan) for DNA bands. The DNA bands were identified according to their molecular size by comparing with the molecular weight marker (100bp DNA ladder) loaded in a separated lane.
2.4.6. Procedure of DNA Sequencing
For sequencing of bacterial DNA, purification of amplified PCR products was done by using DNA purification kits (FAVOGEN, Biotech Corp). Purified PCR products of Proteus mirabilis were sent to 1st Base Laboratories, Malaysia for sequencing by capillary method (ABI PRISM 3500). BLAST analysis was performed to search for homologous sequences into the GenBank at www.ncbi.nlm.nih.gov.
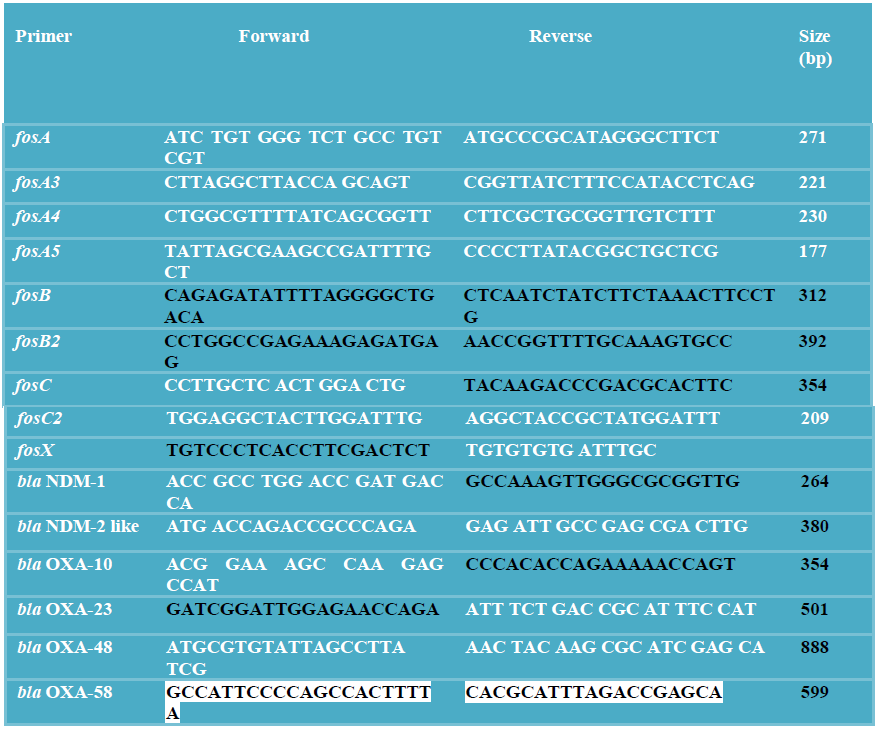
2.4.7. Primers used in this study [20, 5, 4, 28]
2.5. In vivo studies
The experiment was performed in immunocompetent male and female Swiss albino mice weighing 12-15 grams. Mice were infected by intraperitoneal injection of 12.5 units of approximately 104cfu/ml bacterial inoculums using a 100 IU insulin syringe in the lower right abdomen [41]. Bacterial inoculum was obtained through a 24 hours subculture of multidrug resistant Proteus mirabilis in MHA media at 37°C. The animals were observed for 72 hours and the survival rates were recorded every 12 hours. Blood samples were taken as detailed below. All the samples were processed for microbiological studies.
2.5.1. In vivo different antibiotic regimens studies
To evaluate the effectiveness of the different antibiotic regimens, 50 mice were divided into 10 groups with 5 mice in each group. In group A imipenem (dose- 60mg/kg/day, in group B tigecycline (dose-20mg/kg/day), in group C amikacin (dose-15mg/kg/day), in group D imipenem and amikacin, in group E imipenem and tigecycline, in group F tigecycline and amikacin, in group G fosfomycin (dose-400mg/kg/day), in group H fosfomycin and imipenem were injected. Group I was positive control and group J was negative control. Group A, B, C, D, E, F, G, H and I were inoculated with bacterial inoculums. But Group J was not inoculated with bacterial inoculums. Group A, B, C, D, E, F, G, H and J received antimicrobial treatment. Group I was inoculated with bacterial inoculums but did not receive antimicrobial treatment. Fosfomycin was given orally and all other drugs were given intraperitoneally for 72 hours.
The first dose of every antibiotic was administered 4 hours after inoculation of the organism. In order to confirm that these drugs were not toxic to the animal, another group of five uninfected mice (Group J) were given each antibiotic for 72 hours (uninfected treatment group/ negative control). The infected animals were observed for 72 hours of treatment and the cumulative survival rates were recorded every 12 hours. Blood samples were taken as described below.
After 72 hours of antibiotic treatment, blood samples were collected from mice by cardiac puncture aseptically. At first, the upper part of the chest was shaved by razor, washed with hexisol. After palpating the cardiac 61 pulsation with the finger pulp, the area was washed with povidone iodine, 100IU insulin syringe needle was introduced through the skin in the heart of the mouse blindly. For blood culture 1.5 ml of each mouse's blood was collected and then inoculated in sterile conical flask with 5 ml of TSB and incubated for 24 hours at 37ºC. Subculture was done in blood agar and MacConkey agar media and incubated for 24 hours at 37ºC. Then the incubated plates were observed for positive or negative growth. Subculture was done after 48 hours and 72 hours if culture yielded no growth after 24 hours.
3. Results
Among 350 culture positive samples, 244 were wound swab and pus, 84 were urine and 22 were blood samples. Among 244 wound swab and pus samples, 26 (10.65%) were P. mirabilis, among 84 urine samples, 8 (9.52%) were P. mirabilis and among 22 blood samples, 3 (13.64%) were P. mirabilis (Table-I).
Table I: Distribution of P. mirabilis isolated from different culture positive samples (N=350).

N= Number of culture positive samples.
n = Number of isolated P. mirabilis from different culture positive samples.
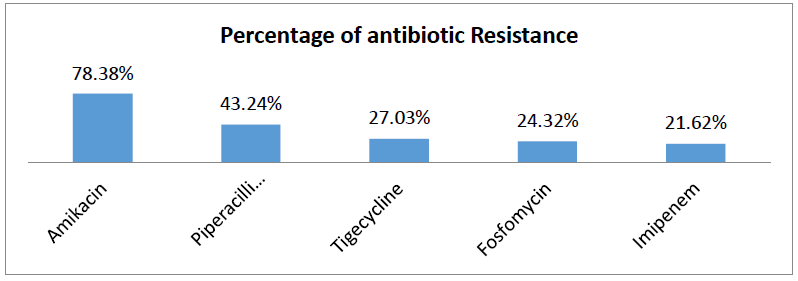
Among 37 isolated Proteus mirabilis, 78.38%, were resistant to amikacin, 43.24% were resistant to piperacillin-tazobactam, 27.03% were resistant to tigecycline, 24.32% were resistant to fosfomycin and 21.62% were resistant to imipenem (Table-II).
Table II: Antibiotic resistance pattern of P. mirabilis.
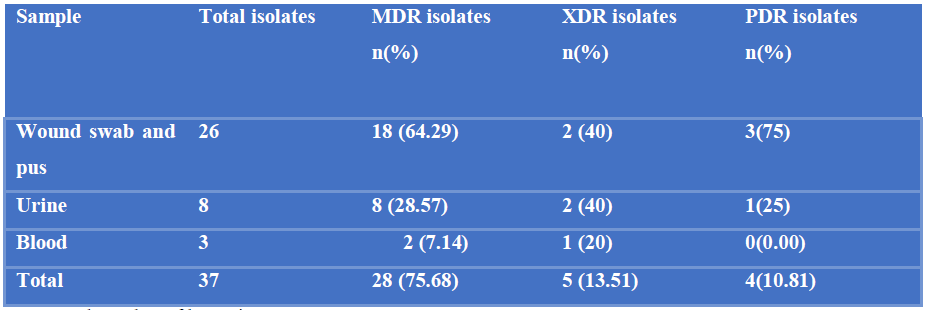
Of the isolated 37 Proteus mirabilis, 28 (75.68%) were MDR of which 18 (64.29%) were detected from wound swab and pus, 8 (28.57%) from urine and 2 (7.14%) from blood samples; 5 (13.51%) were XDR, of which 2 (40%) were detected from wound swab and pus, 2 (40%) from urine and 1 (20%) from blood samples and 4 (10.81%) were PDR, among them 3 (75%) were detected from wound swab and pus and 1 (25%) from urine samples (Table-III).
Table III: Distribution of multidrug resistant (MDR) extensively drug resistant (XDR) and pan drug resistant (PDR) P. mirabilis isolated from different samples.
N= Total number of bacteria.
n= Number of MDR/XDR/PDR P. mirabilis isolated from different samples.
Among 9 fosfomycin resistant P. mirabilis, 6 (66.67%) were from wound swab and pus and 3 (33.33%) were from urine. Among 8 imipenem resistant Proteus mirabilis, 4 (50%) were from wound swab and pus and 4 (50%) were from urine. Among 10 tigecycline resistant Proteus mirabilis, 4 (40%) were from wound swab and pus and 6 (60%) was from urine. Among 28 amikacin resistant P. mirabilis, 17 (60.71%) were from wound swab and pus, 9 (32.14%) were from urine and 2 (7.14%) were from blood (Table-IV).
Table IV: Distribution of fosfomycin resistant, imipenem resistant, tigecycline resistant and amikacin resistant P. mirabilis in different samples.
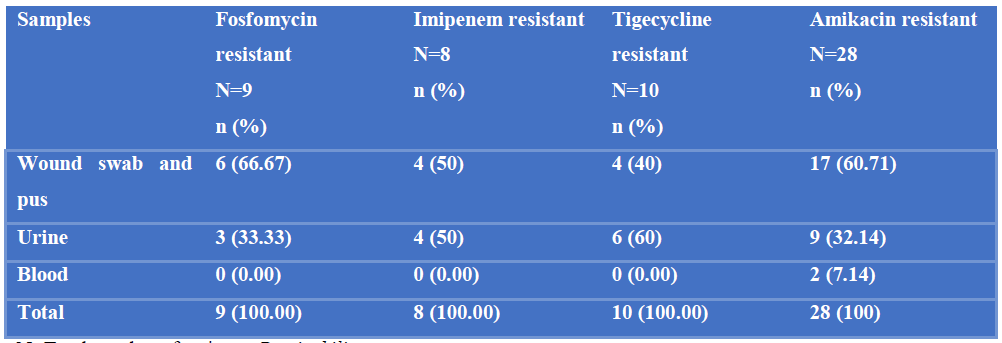
N=Total number of resistant P. mirabilis.
n= Number of antibiotic resistant P. mirabilis from different samples.
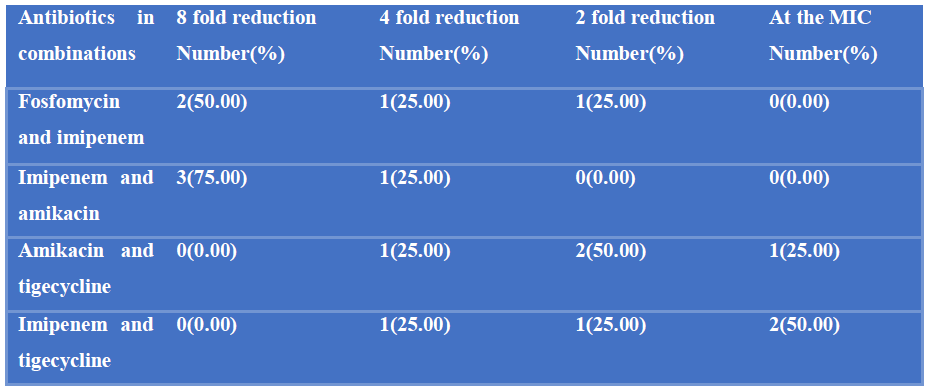
Out of 4 fosfomycin and imipenem resistant P. mirabilis, 2(50%) had 8 fold reduction of MIC, one (25%) had 4 fold reduction of MIC and one (25%) had 2 fold reduction of MIC. Among 4 imipenem and amikacin resistant P. mirabilis, 3(75%) had 8 fold reduction of MIC and one (25%) had 4 fold reduction of MIC; Out of 4 amikacin and tigecycline resistant P. mirabilis, one (25%) had 4 fold reduction of MIC, 2(50%) had 2 fold reduction of MIC and 1(25%) had no difference on MIC and Of the 4 imipenem and tigecycline resistant P.mirabilis, one (25%) had 4 fold reduction of MIC, one (25%) had 2 fold reduction of MIC and 2(50%) had no difference on MIC (Table-V).
Table V: Efficacy of antibiotic combinations against MDR P. mirabilis identified by agar dilution method (N=4).
Out of 4 fosfomycin and imipenem resistant strains, 3 had synergism in combination as their FICI value were 0.5 and one had addition in combination as its FICI value was 1. Of the, 4 imipenem and amikacin resistant strains, all showed synergism in combinations as their FICI value were 0.50. Out of 4 amikacin and tigecycline resistant strains, one had FICI value 0.05(synergistic), 2 had FICI value 1 (additive) and one had FICI value 2 (indifference). Out of 4 imipenem and tigecycline resistant strains, one had FICI value 0.05 (synergistic), one had FICI value 1 (additive) and 2 had FICI value 2 (indifference) (Table-VI).
Table VI: Comparison of efficacy of different antibiotic combinations by FICI formula in MDR P. mirabilis.
|
Antimicrobial Combination |
MIC value by agar dilution method (µg/ml) |
FICa+ FICb |
FICI |
Effects |
Mean FICI |
|
Fosfomycin + Imipenem |
Fosfomycin Imipenem |
0.5 |
|||
|
Alone Combination Alone Combination |
|||||
|
4096 512 64 8 |
0.125+0.125 |
0.25 |
Synergistic |
||
|
1024 128 64 8 |
0.125+0.125 |
0.25 |
Synergistic |
||
|
1024 256 32 8 |
0.25+0.25 |
0.50 |
Synergistic |
||
|
128 64 16 8 |
0.5+0.5 |
1 |
Additive |
||
|
Imipenem + Amikacin |
Imipenem Amikacin |
||||
|
Alone Combination Alone Combination |
|||||
|
64 8 32768 4096 |
0.125+0.125 |
0.25 |
Synergistic |
0.31 |
|
|
64 8 2048 256 |
0.125+0.125 |
0.25 |
Synergistic |
||
|
32 4 1024 128 |
0.125+0.125 |
0.25 |
Synergistic |
||
|
16 4 256 64 |
0.25+0.25 |
0.50 |
Synergistic |
||
|
Amikacin + Tigecycline |
Amikacin Tigecycline |
1.125 |
|||
|
Alone Combination Alone Combination |
|||||
|
32768 8192 128 32 |
0.25+0.25 |
0.50 |
Synergistic |
||
|
2048 1024 64 32 |
0.5+ 0.5 |
1 |
Additive |
||
|
1024 512 32 16 |
0.5+ 0.5 |
1 |
Additive |
||
|
256 256 16 16 |
1+1 |
2 |
Indifference |
||
|
Imipenem + Tigecycline |
Imipenem Tigecycline |
1.375 |
|||
|
Alone Combination Alone Combination |
|||||
|
64 16 128 32 |
0.25+0.25 |
0.50 |
Synergistic |
||
|
64 32 64 32 |
0.5+ 0.5 |
1 |
Additive |
||
|
32 32 32 32 |
1+1 |
2 |
Indifference |
||
|
16 16 16 16 |
1+1 |
2 |
Indifference |
||
Note: FICa is fractional inhibitory concentration of one antibiotic in combination.
FICb is a fractional inhibitory concentration of other antibiotics in combination.
After antibiotic therapy, survival of mice by periodic observation after 12, 24, 36, 48, 60 and 72 hours of infection were recorded. After 48 hours 2 mice from the positive control group and after 72 hours another one died. After 60 hours 4 mice from group B treated with tigecycline alone died. However, the mouse group treated with amikacin alone became profoundly sick with advancement of time in comparison to other groups (Table-VII).
Table VII: Survival rate of mouse after antibiotic therapy found by periodic observation.
|
Time after infection (hour) |
12 |
24 |
36 |
48 |
60 |
72 |
|
Positive Control n(%) |
5(100.00) |
5(100.00) |
5(100.00) |
3(60.00) |
3(60.00) |
2(40.00) |
|
Negative Control n(%) |
5(100.00) |
5(100.00) |
5(100.00) |
5(100.00) |
5(100.00) |
5(100.00) |
|
Imipenem n(%) |
5(100.00) |
5(100.00) |
5(100.00) |
5(100.00) |
5(100.00) |
5(100.00) |
|
Tigecycline n(%) |
5(100.00) |
5(100.00) |
5(100.00) |
5(100.00) |
1(20.00) |
1(20.00) |
|
Amikacin n(%) |
5(100.00) |
5(100.00) |
5(100.00) |
5(100.00) |
5(100.00) |
5(100.00) |
|
Imipenem+ Amikacin n(%) |
5(100.00) |
5(100.00) |
5(100.00) |
5(100.00) |
5(100.00) |
5(100.00) |
|
Imipenem+Tigecycline n(%) |
5(100.00) |
5(100.00) |
5(100.00) |
5(100.00) |
5(100.00) |
5(100.00) |
|
Tigecycline+ Amikacin n(%) |
5(100.00) |
5(100.00) |
5(100.00) |
5(100.00) |
5(100.00) |
5(100.00) |
|
Fosfomycin n(%) |
5(100.00) |
5(100.00) |
5(100.00) |
5(100.00) |
5(100.00) |
5(100.00) |
|
Fosfomycin+imipenem n(%) |
5(100.00) |
5(100.00) |
5(100.00) |
5(100.00) |
5(100.00) |
5(100.00) |
Among the mice treated with different antibiotics, 100% of the imipenem treated group, 80% of the fosfomycin treated group and 60% of the tigecycline treated group and amikacin treated group were culture negative. Among the mice treated with different antibiotic combinations, 100% of the imipenem+amikacin treated group and fosfomycin+imipenem treated group, 80% of the imipenem+tigecycline treated group and 60% of the tigecycline+amikacin treated group were culture negative (Figure-1).
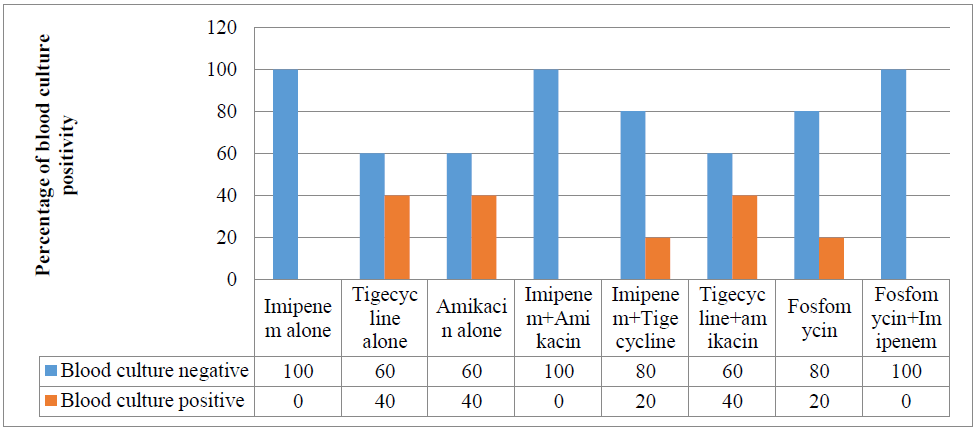
Figure 1: Result of antibiotic therapy on the clearance of MDR P. mirabilis from the blood of mice.
The comparison between synergism of different antibiotic combinations in MDR P.mirabilis in vitro and in vivo was documented (Table-VIII).
Table VIII: Comparison of synergism among different antibiotic combinations between MDR P. mirabilis in vitro and in vivo.

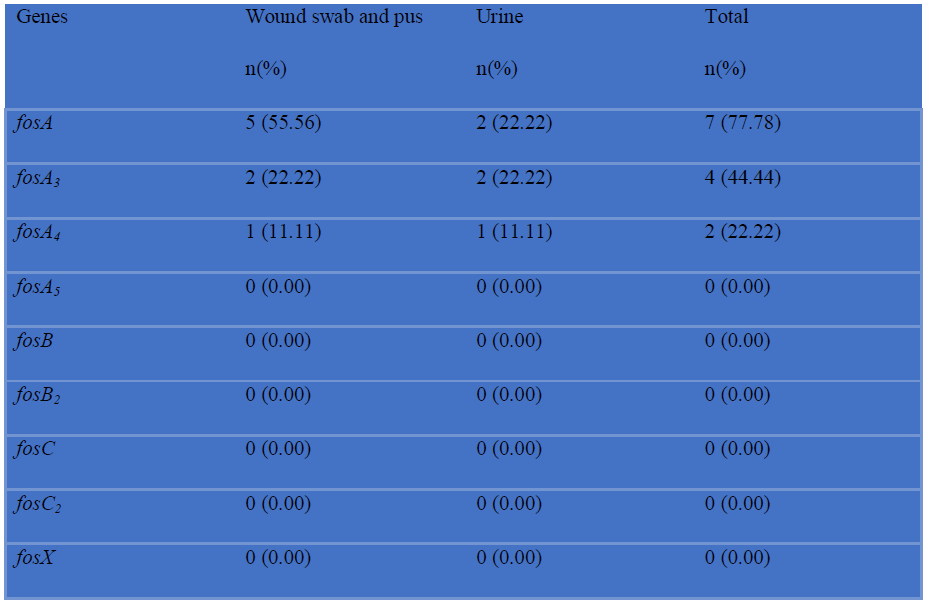
Among 8 imipenem resistant P. mirabilis, 5 (62.50%) were positive for blaNDM-1 and blaOXA-10, 4 (50%) were positive for blaNDM-2, blaOXA-23 and blaOXA-48 and 2 (25%) were positive for blaOXA-58 (Table-IX). Among 9 fosfomycin resistant P. mirabilis, 7 (77.78%) were positive for fosA, 4 (44.44%) were positive for fosA3 and 2 (22.22%) for fosA4. No fosA5, fosB, fosB2, fosC, fosC2 and fosX were detected (Table-X).
Table IX: Detection of blaNDM-1, blaNDM-2 like, blaOXA-10, blaOXA-23, blaOXA-48, blaOXA-58 genes among imipenem resistant P. mirabilis by PCR (N=8).
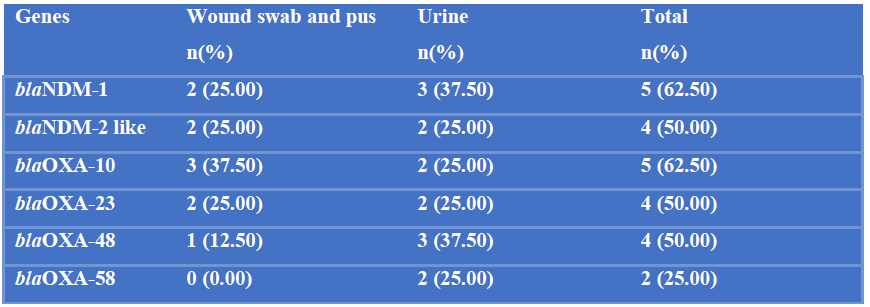
N= Total number of imipenem resistant P. mirabilis.
n= Total number of imipenem resistance gene in different samples.
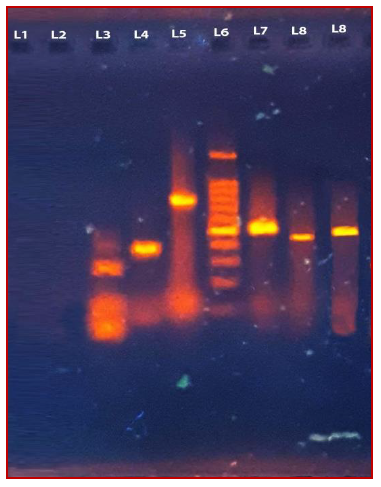
Figure 2: Photograph of gel electrophoresis of amplified DNA of 264bp for NDM-1 gene (lane3), amplified DNA of 354bp for OXA-10 gene (lane 4), amplified DNA of 888bp for OXA-48 gene (lane 5), 1500bp DNA ladder (lane 6), amplified DNA of 599bp for OXA-58 gene (lane 7), amplified DNA of 380bp for NDM-2 like gene (lane 8), amplified DNA of 501bp for OXA-23 gene (lane 9), negative control without DNA (TEbuffer) (lane 1), negative control Escherichia coli ATCC 25922 (lane 2).
Table X: Detection of fosA, fosA3, fosA4, fosA5, fosB, fosB2, fosC, fosC2 and fosX genes among fosfomycin resistant P. mirabilis by PCR (N=9).
Sequencing of fosA, fosA3, fosA4, NDM-1, NDM-2 like, OXA-10, OXA-23 and OXA-58 were done. Of this, NDM-2 like gene got a GenBank accession number and that was MW672265.
4. Discussion
Multidrug resistance is the most important problem in antibiotic resistance due to difficulty in treating multidrug resistant microorganisms and exponential increase in MDR over the last decade [36]. Proteus mirabilis is responsible for a wide range of infections that affect the urinary tract, the respiratory tract, burns, wounds and the feet of individuals with diabetes. They are highly resistant to antimicrobial agents, and new therapeutic options are therefore needed to combat this pathogen [17].
There is limited data regarding Proteus mirabilis as few studies in Bangladesh have been conducted previously. Therefore the present study was designed to find out multidrug resistance among the isolated Proteus mirabilis phenotypically and genotypically with their antibiotic resistance pattern and evaluation of the efficacy of antibiotic combination against MDR Proteus mirabilis both in vitro and in vivo.
24.32% MDR Proteus mirabilis were resistant to fosfomycin. Clinical use of fosfomycin in Bangladesh is rare and there is very little data regarding fosfomycin resistance. The reason behind such finding in present study might be due to horizontal transfer of resistance genes between different species. Recent studies indicate that the recombination of plasmid- encoding carbapenemase and fosfomycinase occurs via mobile elements, presenting new treatment challenges [43].
Resistance to imipenem which is used as a last resort of drugs in the health care settings was found 21.62% in P. mirabilis. P. mirabilis isolates present an elevation in the resistance level to imipenem due to many reasons: the loss of outer membrane porins, decreased expression of PBP1a or reduced binding of imipenem by PBP2 [16]. ImpR (outer membrane protein) over expression resulted in increased carbapenem MICs in the laboratory strain and clinical isolates. Development of resistance against imipenem in P. mirabilis is also due to the absence of 24 kDa OMP [42].
Though colistin and tigecycline are considered as the most sensitive drugs against CRE but P. mirabilis is intrinsically resistant to tigecycline and colistin [25]. 27.03% P. mirabilis were found resistant to tigecycline. For tigecycline, the genome of P. mirabilis contains a homolog of the Esch. coli AcrAB efflux system and this gene cluster appears to be responsible, at least in part, for the intrinsic reduced susceptibility to tigecycline in P. mirabilis [44].
Multidrug resistance to P. mirabilis could be a result of continuous use of broad spectrum antimicrobials and non adherence to hospital antibiotic therapy, horizontal transfer of drug resistance genes among different bacteria as most patients already have been harboring the resistant organisms in their hospital staying period [33].
The present study observed 100% synergism with the combination of imipenem plus amikacin, 75% with fosfomycin plus imipenem and 25% with amikacin plus tigecycline and imipenem plus tigecycline. Fosfomycin in combination with either amino glycosides and carbapenems or piperacillin-tazobactam is the effective combination against MDR Enterobacteriaceae [15]. As with infections with gram-negative bacteria, antimicrobial synergy has traditionally been seen with β-lactam–aminoglycoside combinations. β-lactam mediated disturbance of the cell walls of gram-negative bacilli facilitates passage of amino glycosides into the periplasmic space [27]. Interestingly, time–kill assays disclosed a bactericidal synergism between tigecycline and amikacin against P. vulgaris. Such combinations could be of clinical relevance as the organisms they target belong to potentially problematic multi resistant species and to bacteria inadequately inhibited by tigecycline [12].
In the present study, mice were observed periodically for survival for 72 hours after intervention. Mice of the positive control group became profoundly sick and after 48 hours 2 mice had died and one more died after 72 hours. 4 mice of the tigecycline alone group died after 60 hours. In animal studies, a common outcome of therapy with tigecycline in combination was prolonged survival and/or reductions in tissue colony counts [12].
In vivo combination of imipenem plus amikacin and imipenem plus fosfomycin showed 100% synergism, imipenem plus tigecycline showed 80% synergism and amikacin and tigecycline showed 60% synergism. Fosfomycin is reported to mitigate in vivo and in vitro synergy with carbapenem against KPC producing multidrug resistant Enterobacteriaceae [40, 32]. The best in vivo result appeared in the group treated with imipenem combinations.
Among imipenem resistant P. mirabilis, 62.50% and 50% were positive for blaNDM-1 and blaNDM-2 like genes detected by PCR. In Bangladesh, blaNDM-1 was first identified in K. pneumoniae in 2008 thereafter some reports described detection of blaNDM-1 in some GNR species, [13, 4]. Since the first identification of blaNDM-1 in 2008, worldwide attention was attracted to this carbapenemase because of its rapid dissemination among Enterobacteriaceae and Acinetobacter spp. clinical isolates, as well as those colonizing humans and contaminating environments [10].
Among imipenem resistant P. mirabilis 62.50% were positive for blaOXA-10, 50% were positive for blaOXA-23 and blaOXA-48 and 25% were positive for blaOXA-58. It was found, blaOXA-10was horizontally transferable inEnterobacteriaceaefamily which was supported by transformation and conjugation [26]. Considering the transferable elements surrounding blaOXA-23 as well as the phenotype of relative susceptibility of this isolate, blaOXA genes may be spreading silently “under the radar” [23]. The blaOXA-48 gene is relatively common in Europe, especially in Mediterranean countries [35]. In a study from Turkey, 92% of carbapenem resistant Enterobacteriaceae were OXA-48 like producers [3]. In addition to Europe, OXA-48-like enzymes have been found worldwide in Enterobacteriaceae [31]. The blaOXA-58 represents one of the class D carbapenemase types that are major carbapenem resistance determinants in Acinetobacter spp. being extremely rare in Enterobacteriaceae. The first report of its presence in diverse enterobacteria was from Sierra Leone, followed by recent papers on Proteus mirabilis from Belgium and Germany. Only 6 such isolates, exclusively P. mirabilis, have been reported in Europe to date, of which German and Polish isolates shared unique plasmids [8].
Among the fosfomycin resistant P. mirabilis, 77.78% were positive for fosA, 44.44% for fosA3 and 22.22% were positive for fosA4 genes. The fosfomycin resistance protein FosA, are metalloenzymes that catalyse the nucleophilic addition of the tripeptide glutathione to the C1 position of the antibiotic, cleaving the epoxide ring and rendering it ineffective as an antibacterial drug. Among glutathione transferase (FosA type) enzymes found to be plasmid- borne are FosA3, FosA4, FosA5 and FosC2 [14].
DNA sequence of amplified PCR product and translated nucleotide base sequence of fosA, fosA3 and fosA4 showed point mutations including base substitution and frameshift mutation at multiple positions. Large and small deletions are the main source of gene-inactivating mutations followed by insertions/duplications [19]. In addition a considerable number of point mutations had been detected, including truncation by nonsense and missense mutations [37].
5. Conclusion
From in vivo and in vitro experiments, it may be clear that imipenem and amikacin is the best combination for MDR P. mirabilis. The second best in vivo and in vitro effective combination is imipenem and fosfomycin.
Repurposing of older antimicrobials like fosfomycin and imipenem combination therapy may be good options for the treatment of infection caused by them.
6. References
- Andrews JM. Determination of minimum inhibitory concentrations. Journal of antimicrobial Chemotherapy 48 (2001): 5-16.
- Aris P, Boroumand MA, Rahbar M, Douraghi M. The activity of fosfomycin against extended-spectrum beta-lactamase-producing isolates of Enterobacteriaceae recovered from urinary tract infections: a single-center study over a period of 12 years.Microbial Drug Resistance 24(2018): 607-612.
- Baran I, Aksu N. Phenotypic and genotypic characteristics of carbapenem-resistant Enterobacteriaceae in a tertiary-level reference hospital in Turkey.Annals of clinical microbiology and antimicrobials 15(2016): 20.
- Begum N, Shamsuzzaman SM. Emergence of carbapenemase-producing urinary isolates at a tertiary care hospital in Dhaka, Bangladesh. Tzu Chi Medical Journal 28 (2016): 94-98.
- Benzerara Y, Gallah S, Hommeril B, Genel N, Decré D, Rottman M, Arlet G. Emergence of plasmid-mediated fosfomycin-resistance genes among Escherichia coli isolates, France. Emerging infectious diseases 23 (2017): 1564.
- Charan J, Mulla S, Ryavanki S, Kantharia N. New Delhi Metallo–beta lactamase–1 containing Enterobacteriaceae: origin, diagnosis, treatment and public health concern. Pan African Medical Journal 11 (2012).
- Cheesbrough M. Antimicrobial susceptibility testing. In: Cheesbrough M, (editor). District Laboratory Practice in Tropical countries, part 2, 2nd ed. Cambridge University press (2009):132-143.
- Chen TL, Chang WC, Kuo SC, Lee YT, Chen CP, Siu LK, Cho WL, Fung CP. Contribution of a plasmid-borne blaOXA-58 gene with its hybrid promoter provided by IS1006 and an ISAba3-like element to β-lactam resistance in Acinetobacter genomic species 13TU. Antimicrobial agents and chemotherapy 54 (2010):3107-12.
- Clinical and Laboratory Standard Institute (CLSI) performance standards for susceptibility testing; Twenty-ninth informational supplement. CLSI document M100- S28 (2019).
- Dortet L, Poirel L, Nordmann P. Worldwide dissemination of the NDM-type carbapenemases in Gram-negative bacteria. BioMed research international (2014).
- Eliopoulos GM, Eliopoulos CT. Antibiotic combinations: should they be tested? Clinical Microbiology Reviews 1 (1988):139-56.
- Entenza JM, Moreillon P. Tigecycline in combination with other antimicrobials: a review of in vitro, animal and case report studies. International journal of antimicrobial agents 34 (2009): 8-e1.
- Farzana R, Shamsuzzaman SM, Mamun KZ. Isolation and molecular characterization of New Delhi metallo-beta-lactamase-1 producing superbugs in Bangladesh. The Journal of Infection in Developing Countries 7 (2013):161-8.
- Fillgrove KL, Pakhomova S, Newcomer ME, Armstrong RN. Mechanistic diversity of fosfomycin resistance in pathogenic microorganisms. Journal of the American Chemical Society 125 (2003): 15730-1.
- Giancola SE, Mahoney MV, Hogan MD, Raux BR, McCoy C, Hirsch EB. Assessment of fosfomycin for complicated or multidrug-resistant urinary tract infections: patient characteristics and outcomes. Chemotherapy 62 (2017):100-4.
- Girlich D, Dortet L, Poirel L, Nordmann P. Integration of the bla NDM-1 carbapenemase gene into Proteus genomic island 1 (PGI1-Pm PEL) in a Proteus mirabilis clinical isolate. Journal of Antimicrobial Chemotherapy 70 (2015): 98-102.
- Gomaa S, Serry F, Abdellatif H, Abbas H. Elimination of multidrug-resistant Proteus mirabilis biofilms using bacteriophages. Archives of virology 164 (2019): 2265-75.
- Gombert ME, Aulicino TM. Synergism of imipenem and amikacin in combination with other antibiotics against Nocardia asteroides. Antimicrobial agents and chemotherapy 24 (1983): 810-1.
- Gracia-Aznarez FJ, Fernandez V, Pita G, Peterlongo P, Dominguez O, de la Hoya M, Duran M, Osorio A, Moreno L, Gonzalez-Neira A, Rosa-Rosa JM. Whole exome sequencing suggests much of non-BRCA1/BRCA2 familial breast cancer is due to moderate and low penetrance susceptibility alleles. PloS one 8 (2013).
- Ho PL, Chan J, Lo WU, Lai EL, Cheung YY, Lau TC, Chow KH. Prevalence and molecular epidemiology of plasmid-mediated fosfomycin resistance genes among blood and urinary Escherichia coli isolates. Journal of medical microbiology 62 (2013):1707-13.
- Islam TA, Shamsuzzaman SM, Farzana A. Prevalence and anti biogram of ESBL producing gram negative bacilli isolated from urine in Dhaka Medical College Hospital, Bangladesh. Bangladesh Journal of Medical Microbiology 9 (2015): 17-21.
- Kukavica-Ibrulj I. and Levesque RC. Animal models of chronic lung infection with Pseudomonas aeruginosa: useful tools for cystic fibrosis studies.Laboratory animals42 (2008): 389-412.
- La MV, Jureen R, Lin RT, Teo JW. Unusual detection of an Acinetobacter class D carbapenemase gene, bla OXA-23, in a clinical Escherichia coli isolate. Journal of clinical microbiology 52 (2014):3822-3.
- Lambert RJW, Johnston MD, Hanlon GW, Denyer SP. Theory of antimicrobial combinations: biocide mixtures–synergy or addition?Journal of applied microbiology94 (2003): 747-759.
- Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Flagas ME, Giske CG et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria. Clin Microbiol Infect 18 (2012): 268-281.
- Maurya AP, Dhar D, Basumatary MK, Paul D, Ingti B, Choudhury D, Talukdar AD, Chakravarty A, Mishra S, Bhattacharjee A. Expansion of highly stable bla OXA-10 β-lactamase family within diverse host range among nosocomial isolates of Gram-negative bacilli within a tertiary referral hospital of Northeast India. BMC research notes 10 (2017):145.
- Miller MH, Feinstein SA, Chow RT. Early effects of beta-lactams on aminoglycoside uptake, bactericidal rates, and turbidimetrically measured growth inhibition in Pseudomonas aeruginosa. Antimicrobial agents and chemotherapy 31 (1987):108-10.
- Mlynarcik P, Roderova M, Kolar M. Primer Evaluation for PCR and its Application for Detection of Carbapenemases in Enterobacteriaceae. Jundishapur journal of microbiology 9 (2016).
- Nakamura G, Wachino JI, Sato N, Kimura K, Yamada K, Jin W, Shibayama K, Yagi T, Kawamura K, Arakawa Y. Practical agar-based disk potentiation test for detection of fosfomycin-nonsusceptible Escherichia coli clinical isolates producing glutathione S-transferases.Journal of clinical microbiology52 (2014):3175-3179.
- Nordmann P, Naas T, Poirel L. Global spread of carbapenemase-producing Enterobacteriaceae. Emerging infectious diseases 17 (2011): 1791.
- Ocampo AM, Chen L, Cienfuegos AV, Roncancio G, Chavda KD, Kreiswirth BN, Jiménez JN. A two-year surveillance in five Colombian tertiary care hospitals reveals high frequency of non-CG258 clones of carbapenem-resistant Klebsiella pneumoniae with distinct clinical characteristics. Antimicrobial agents and chemotherapy 60 (2016): 332-42.
- Perdigão Neto LV, Oliveira MS, Martins RC, Marchi AP, Gaudereto JJ, da Costa LA, de Lima LF, Takeda CF, Costa SF, Levin AS. Fosfomycin in severe infections due to genetically distinct pan-drug-resistant Gram-negative microorganisms: synergy with meropenem. Journal of Antimicrobial Chemotherapy 74 (2019): 177-81.
- Perween N, Prakash SK, Bharara T. Prevalence of Multidrug-Resistant and Extensively Drug-Resistant Proteus, Providencia and Morganella Species in Burn Wound Infection. Int J Sci Stud 3 (2016):154-156.
- Pilato D, Chiarelli V, Boinett A, Riccobono CJ, Harris E, D'Andrea SR, Thomson MM, Rossolini NR, Giani T. Complete genome sequence of the first KPC-type carbapenemase-positive Proteus mirabilis strain from a bloodstream infection.Genome Announc4 (2016): 00607-16.
- Poirel L, Potron A, Nordmann P. OXA-48-like carbapenemases: the phantom menace. Journal of Antimicrobial Chemotherapy 67 (2012): 1597-606.
- Ramos JR, Lletí MS. Fosfomycin in infections caused by multidrug-resistant Gram-negative pathogens. Revista Española de Quimioterapia 32 (2019):45.
- Rodríguez-Hernández MJ, Pachón J, Pichardo C, Cuberos L, Ibáñez-Martínez J, García-Curiel A, Caballero FJ, Moreno I, Jiménez-Mejías ME. Imipenem, doxycycline and amikacin in monotherapy and in combination in Acinetobacter baumannii experimental pneumonia. Journal of Antimicrobial Chemotherapy 45 (2000):493-501.
- Saiprasad PV, Krishnaprasad K. Exploring the hidden potential of fosfomycin for the fight against severe Gram-negative infections.Indian journal of medical microbiology 34 (2016):416.
- Sopirala MM, Mangino JE, Gebreyes WA, Biller B, Bannerman T, Balada-Llasat JM, Pancholi P. Synergy testing by Etest, microdilution checkerboard, and time-kill methods for pan-drug-resistant Acinetobacter baumannii. Antimicrobial agents and chemotherapy 54 (2010): 4678-83.
- Souli M, Galani I, Boukovalas S, Gourgoulis MG, Chryssouli Z, Kanellakopoulou K, Panagea T, Giamarellou H. In vitro interactions of antimicrobial combinations with fosfomycin against KPC-2 producing Klebsiella pneumoniae and protection of resistance development. Antimicrobial agents and chemotherapy 55 (2011): 2395-7.
- Toledo AVM,Tuon FF,Bail L, Manente F, Arruda P, Aranha-Junior AA. Experimental model for treatment of extended spectrum beta lactamase producing-klebsiella pneumonia. Arq Bras Cir Diag 27 (2014): 168-171.
- Tsai YL, Wang MC, Hsueh PR, Liu MC, Hu RM, Wu YJ, Liaw SJ. Overexpression of an outer membrane protein associated with decreased susceptibility to carbapenems in Proteus mirabilis. PloS one 10 (2015).
- Tseng SP, Wang SF, Ma L, Wang TY, Yang TY, Siu LK, Chuang YC, Lee PS, Wang JT, Wu TL, Lin JC. The plasmid-mediated fosfomycin resistance determinants and synergy of fosfomycin and meropenem in carbapenem-resistant Klebsiella pneumoniae isolates in Taiwan. Journal of microbiology, immunology and infection 50 (2017): 653-61.
- Visalli MA, Murphy E, Projan SJ, Bradford PA. AcrAB multidrug efflux pump is associated with reduced levels of susceptibility to tigecycline (GAR-936) in Proteus mirabilis. Antimicrobial agents and chemotherapy 47 (2003): 665-9.
- World Health Organization. WHO Advisory Group on Integrated Surveillance of Antimicrobial Resistance (AGISAR). Critically important antimicrobials for human medicine 3rd Revision 2011. WHO Document Production Services, Geneva, Switzerland. Clin. Infect, Dis 55 (2012): 712-9.
- Xu H, Miao V, Kwong W, Xia R, Davies J. Identification of a novel fosfomycin resistance gene (fosA2) in Enterobacter cloacae from the Salmon River, Canada. Letters in applied microbiology 52 (2011): 427-9.


 Impact Factor: * 3.5
Impact Factor: * 3.5 Acceptance Rate: 71.36%
Acceptance Rate: 71.36%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 
