Probiotic potential of Lactobacilli and Bifidobacteria found in breast milk in Gabon: Isolation, Identification, Antibacterial activity and Perspective for Antibiotherapy
Natacha Moussadji1,2, Brice Boris Legba2, Kossiwa Kokou1, Paulin N Essone1, Vanessa Amana-Bokagne1,4, Edlom Pélagie Tchadie1,4, Lamine Saïd Baba-Moussa3, Victorien Dougnon2, Selidji Todagbé Agnandji1,4,5 And Gatien Lokossou*,1
1Centre de Recherches Médicales de Lambaréné, Lambaréné, Gabon.
2Research Unit in Applied Microbiology and Pharmacology of natural substances (URMAPha); Research Laboratory in Applied Biology, Polytechnic School of Abomey-Calavi, University of Abomey-Calavi. Benin
3Laboratory of Biology and Molecular Typing in Microbiology, Faculty of Sciences and Technology, University of Abomey-Calavi, Benin
4Institut für Tropenmedizin, Universität Tübingen, Tübingen, Germany
5Institute of Medical Microbiology, University Hospital Münster, Münster, Germany
*Corresponding author: Gatien Lokossou, Centre de Recherches Médicales de Lambaréné, Lambaréné, Gabon.
Received: 05 July 2025; Accepted: 23 August 2025; Published: 03 October 2025
Article Information
Citation: Natacha Moussadji, Brice Boris Legba, Kossiwa Kokou, Paulin N Essone, Vanessa Amana- Bokagne, Edlom Pélagie Tchadie, Lamine Saïd Baba-Moussa, Victorien Dougnon, Selidji Todagbé Agnandji and Gatien Lokossou. Probiotic potential of Lactobacilli and Bifidobacteria found in breast milk in Gabon: Isolation, Identification, Antibacterial activity and Perspective for Antibiotherapy. Archives of Microbiology and Immunology. 9 (2025): 235-247.
View / Download Pdf Share at FacebookAbstract
Breast milk is considered the optimal source of nutrition for infants due to its rich bioactive components, including probiotic lactic acid bacteria. These bacteria contribute to gut microbiota balance and prevent the colonization of pathogenic microorganisms. This study aimed to evaluate the probiotic potential of lactic acid bacteria isolated from breast milk in Gabon, with a focus on their antibacterial properties and possible applications in antibiotic therapy. Thirty-four breast milk samples, including colostrum, transitional, and mature milk, were collected from lactating women in Gabon. The bacterial strains were isolated and identified using biochemical and molecular techniques. Their antibiotic susceptibility was assessed using the diffusion method. The probiotic potential of the strains was explored through their ability to survive in simulated gastric and intestinal conditions, growth under varying temperatures, and antibacterial activity against clinically relevant pathogens such as Salmonella typhimurium, Staphylococcus aureus, and Escherichia coli (ATCC 25922). A total of 49 bacterial strains were isolated, with Lactobacillus acidophilus being the most prevalent species across all milk types. The strains exhibited significant resistance to acidic and bile conditions, indicating their capacity to colonize the gut effectively. Additionally, they demonstrated strong antibacterial activity, highlighting their potential role in controlling gastrointestinal infections. The presence of such probiotic strains in Gabonese breast milk underscores the importance of breastfeeding in infant health, particularly in low-income populations where access to medical care is limited. These findings suggest that lactic acid bacteria from breast milk could serve as a natural source of probiotics with antimicrobial properties. Future research should focus on characterizing their bioactive compounds and exploring their application in probiotic supplements or therapeutic formulations for combating bacterial infections.
Keywords
<p>Breast milk; Lactobacilli and Bifidobacteria; Probiotic specific functionalities; Gabon</p>
Article Details
Introduction
Probiotics are defined by the Food and Agriculture Organization (FAO) and the World Health Organization (WHO) as “live microorganisms which, when administered in adequate amounts, confer a health benefit on the host.” These beneficial microbes, primarily from the genera Lactobacillus and Bifidobacterium, are known for their diverse clinical and immunological properties. Their effects are strain-specific, meaning that a rational selection of probiotic candidates is essential for targeted therapeutic applications (Jamyuang et al., 2019). The selection of probiotic strains from appropriate sources, such as newborns, children, pregnant women, or the elderly, whose gut microbiota differs from that of healthy adults, is a promising approach for improving health outcomes (Łubiech and Twarużek, 2020).
A recent study reported a reduction in the incidence of infectious diseases in breastfed infants compared to those receiving other dairy products (Juharji et al., 2022). This indicates that specific factors are present in breast milk that appear to provide protection against infectious diseases. Breastfeeding is an important source of lactic acid bacteria for the infant, which can also protect the infant from pathogenic microbes (Liu et al., 2020). Breast milk is a highly nutritious and biologically active fluid that provides essential macronutrients, micronutrients, and bioactive compounds, including antibodies, hormones, growth factors, and antioxidants. Increasing evidence suggests that breast milk plays a crucial role in shaping the neonatal gut microbiota and strengthening the immune system, thus reducing the incidence of infectious diseases in breastfed infants compared to those fed with formula milk (Rodríguez Arreola et al., 2020), along with protective factors (Lokossou et al., 2022).
Increasing evidence has revealed that altered neonatal colonization and disrupted interactions between gut microbes and the host during the neonatal period, especially during breastfeeding (content of the milk), could affect health later in life. Among the bioactive components of breast milk, lactic acid bacteria, particularly Lactobacillus and Bifidobacterium species, have been recognized for their probiotic properties. These bacteria contribute to gut homeostasis by inhibiting the colonization of pathogenic microbes through competitive exclusion, the production of bacteriocins, organic acids, and hydrogen peroxide (Lyons et al., 2020; Javed et al., 2021). These bacteria can also play a significant role in the incidence and severity of infections in breastfed infants. Specific strains of probiotic lactic acid bacteria isolated from breast milk have demonstrated their ability to inhibit the growth of a wide range of pathogenic bacteria through competitive exclusion or by producing antimicrobial components, such as bacteriocins and organic acids (Gunyakti and Asan-Ozusaglam, 2019). It is estimated that breastfed infants receive 104 to 106 bacteria per day (based on an average daily consumption of 800 ml of milk), with most isolated species belonging to the genera Staphylococcus, Streptococcus, Lactobacillus et Bifidobacterium spp. (Sakwinska et al., 2016; Selvamani et al., 2021).
Infants fed with formula milk are not exposed to these potentially beneficial bacteria for health, which is why the provision of Bifidobacterium and Lactobacillus spp. probiotics in infant formulas or milk substitutes remains a priority (Chong et al., 2022). The diversity of probiotic lactic acid bacteria isolated from breast milk and their characteristics have been studied and published (Moossavi and Azad, 2020). Isolating probiotic bacteria from breast milk (exerting its protective effect via the production of hydrogen peroxide and organic acid) is an attractive approach for selecting probiotic strains, as their in vivo production might play a significant role in preventing infectious diseases in the host's gastrointestinal tract (Mohammadi et al., 2018).
Several studies have explored the probiotic potential of LAB isolated from human sources, including the feces of adults and children, based on their ability to withstand bile salts, low pH, and lysozyme, as well as their adhesion capacity to intestinal cells and immunomodulatory properties(Shokryazdan et al., 2017; Amadoro et al., 2018). Additionally, numerous factors influence the composition of breast milk, including maternal age, lactation stage, body mass index (BMI), dietary habits, smoking, parity, place of residence, and environmental conditions (Zacarías et al., 2011; Riaz Rajoka et al., 2017; Nikolopoulou et al., 2021; D’Alessandro et al., 2022). While extensive research has been conducted on the microbiota of breast milk in different regions worldwide, data on the composition and probiotic potential of LAB in breast milk from African populations, particularly in Gabon, remain scarce. The only existing study on breast milk composition in Gabon, conducted by Essomo et al. (2013), focused primarily on macronutrient content, such as lipids, carbohydrates, proteins, and minerals, without investigating the microbial profile or probiotic potential of the bacteria present. Therefore, the existing limitations in our understanding of the Gabonese breast milk microbiome need to be documented.
Given this knowledge gap, it is essential to characterize and evaluate the probiotic potential of Lactobacillus and Bifidobacterium strains isolated from the breast milk of healthy breastfeeding women in Gabon. These bacteria could have significant implications for infant health by serving as natural probiotics capable of enhancing gut microbiota balance and providing antibacterial activity against common pathogenic bacteria. This study aims to isolate, identify, and assess the antibacterial activity of lactic acid bacteria strains found in breast milk in Gabon, with a perspective toward developing probiotic supplements for infants and exploring their potential applications in antibiotic therapy. The research was conducted in Port-Gentil, Gabon, with institutional approval from the Centre de Recherches Médicales de Lambaréné, marking the first study of its kind in this region. By filling this gap in knowledge, this study seeks to contribute to the growing body of research on breast milk microbiota and its implications for neonatal health. The identification of promising probiotic strains could pave the way for new therapeutic applications, including the formulation of probiotic-based interventions to support infant health and combat bacterial infections more effectively.
Material and methods
Biological materials
This study involved 34 breast milk samples collected from lactating women in Gabon. The samples were classified into colostrum, transitional milk, and mature milk. Pathogenic strains used for antibacterial testing included Escherichia coli ATCC 25299, Salmonella typhimurium, and Staphylococcus aureus, which were obtained from clinical isolates.
Inclusion criteria
Healthy, breastfeeding Gabonese women who provided informed consent participated in the study. They were not undergoing any medical treatments at the time of the research and had no history of antibiotic use for at least six months before and after delivery. This criterion was set to minimize the impact of antibiotics on the milk microbiota. Before their participation, a detailed discussion about the research objectives, the procedures involved, and the potential implications was conducted to ensure they fully understood and voluntarily agreed to take part in the study. This has ensured compliance with ethical standards. Pregnancy-related medical history and previous illnesses were also recorded 6 months before and 6 months after delivery.
Ethics approval statement
The study was approved by the Ethical Committee of the Centre de Recherches Médicales de Lambaréné under the code: CEI-CERMEL/006/2022.
Methods
Sample collection and bacterial isolation
A detailed questionnaire was administered to collect sociodemographic data, medical history, and lifestyle factors influencing breast milk composition. The biological samples consisted of three types of milk: colostrum, transitional milk, and mature milk. Breast milk samples were collected at the maternal and child health center in Port-Gentil. The nipple and areola were disinfected with 70% ethanol before collection. The first few drops were discarded, and subsequent milk was collected in sterile glass tubes. Samples were transported to the laboratory within 24 hours under controlled temperature conditions.
The isolation process was conducted following Liu et al. (2020) with modifications to optimize the recovery of Lactobacillus and Bifidobacterium strains from breast milk (Liu et al., 2020). Two enrichment solutions were prepared per sample: SMs1, consisting of 5 mL of breast milk mixed with 20 mL of sterile peptone water, and SMs2, composed of 5 mL of breast milk supplemented with 20 mL of sterile peptone water and 0.05% cysteine-HCl to create an anaerobic environment favorable for Bifidobacterium growth. Both solutions were incubated anaerobically at 37°C for 24 hours to mimic the natural physiological conditions of breast milk within the human body and facilitate optimal bacterial growth. After incubation, serial dilutions (10-1 to 10-4) were performed, and aliquots from SMs1 were plated onto Man Rogosa Sharp (MRS) agar for Lactobacillus isolation, while SMs2 dilutions were plated onto MRS agar supplemented with 0.05% cysteine-HCl to promote the selective growth of Bifidobacterium species. Plates were incubated anaerobically at 37°C for 24-48 hours, ensuring the recovery of strains capable of thriving under human body temperature. The rationale for additional temperature tests at 15°C and 30°C was to assess the adaptability of the isolated strains to different environmental conditions, which is relevant for their potential probiotic applications, storage stability, and industrial processing. Following incubation, bacterial colonies were purified through three successive subcultures to ensure strain purity and subsequently stored in glycerol at -20°C for long-term preservation and further analysis (Hadadji, 2007).
Molecular identification
The genomic DNA extraction from isolated from colony of each lactic acid bacteria strain recovered on nutrient agar platted culture. It was extracted in accordance with the instructions for the Zymo Quick-DNA Miniprep Plus extraction kit. The optical density was checked by spectrophotometry between 200 and 4000nm to ensure the integrity of the extracted DNA. Once the DNA had been extracted, the specific genes of each bacterial species were analyzed using polymerase chain reaction (PCR) (D’Alessandro et al., 2022). Simplex PCR amplification was carried out using primers targeting genes coding for 16s and 23s rRNAs and their intergenic spacer region, namely R16 and LbLMA1-rev. The PCR conditions were set as follows: initial denaturation at 95°C for 5min, followed by 30 cycles of denaturation at 95°C for 30s, hybridization at 55°C for 30s, extension at 72°C for 30s and a final extension step for 7 min at 72°C. The PCR products were analysed on a 1% agarose gel with a 100 base pair DNA ladder (Takara ,3407A). The primers used for the amplification are listed in table 1 (Liu et al., 2020; Nikolopoulou et al., 2021).
Table 1: List of primers used for lactic acid bacteria identification
|
Bacterial species |
Primers |
Sequences (5’ to 3’) |
Size (pb) |
|
All Lactobacillus |
IDL03R |
CCACCTTCCTCCGGTTTGTCA |
- |
|
All Lactobacillus |
IDL04F |
AGGGTGAAGTCGTAACAAGTAGCC |
- |
|
Lactobacillus casei |
IDL11F |
TGGTCGGCAGAGTAACTGTTGTCG |
727 |
|
Lactobacillus acidophilus |
IDL22R |
AACTATCGCTTACGCTACCACTTTGC |
606 |
|
Lactobacillus delbrueckii |
IDL31F |
CTGTGCTACACCTAGAGATAGGTGG |
184 |
|
Lactobacillus gasseri |
IDL42R |
ATTTCAAGTTGAGTCTCTCTCTC |
272 |
|
Lactobacillus reuteri |
IDL52F |
ACCTGATTGACGATGGATCACCAGT |
1105 |
|
Lactobacillus plantarum |
IDL62R |
CTAGTGGTAACAGTTGATTAAAACTGC |
428 |
|
Lactobacillus rhamnosus |
IDL73R |
GCCAACAAGCTATGTGTTCGCTTGC |
448 |
|
Bifidobacterium |
lm26-f |
GATTCTGGCTCAGGATGAACG |
- |
|
Bifidobacterium |
lm3-r |
CGGGTGCTCCCACTTTCATG |
- |
Antibiotics susceptibility testing
The antibiotic resistance profile of the isolated Lactobacillus and Bifidobacterium strains was assessed using the disk diffusion method, following the guidelines established by the Comité de l'Antibiogramme de la Société Française de Microbiologie (CASFM, 2022). A panel of antibiotics was selected to evaluate the susceptibility of the strains, including Penicillin (10U/L), Ampicillin (10µg), Oxacillin (1µg), Cefoxitin (30µg), Vancomycin (10µg), Erythromycin (15µg), Tetracycline (30µg), Clindamycin (2µg), Chloramphenicol (30µg), Streptomycin (10µg), Gentamicin (10µg), Nalidixic acid (30µg), and Trimethoprim (5µg). To ensure consistency in bacterial inoculum, bacterial suspensions were adjusted to a 0.5 McFarland standard, corresponding to an optical density of approximately 1.5 × 108 CFU/mL. The standardized suspensions were then spread evenly onto MRS agar plates, a medium optimized for the growth of lactic acid bacteria, ensuring optimal growth conditions for the strains. Antibiotic disks were carefully placed on the inoculated plates, which were then incubated anaerobically at 37°C for 24 hours to mimic physiological conditions and allow for proper bacterial growth. Following incubation, the diameters of the inhibition zones surrounding each antibiotic disk were measured in millimeters, and the results were interpreted according to CASFM guidelines, classifying the strains as susceptible, intermediate, or resistant based on established clinical breakpoints. This evaluation provided critical insights into the antibiotic susceptibility patterns of the isolated probiotic strains, helping to determine their potential safety for therapeutic applications and their resistance to commonly used antibiotics. E. coli ATCC 25922 strain was used as quality control.
In vitro probiotic capacity of strains
- • Antimicrobial activity
The antibacterial activity of Lactobacillus and Bifidobacterium strains was tested against multi-resistant isolated at the medical laboratory of the Clinique des Assalas in Port-Gentil/Gabon (E. coli ATCC 25922, S. aureus, and S. typhimurium) (Agbankpe et al., 2019). Briefly, isolated Lactobacillus and Bifidobacterium strains were individually inoculated onto MRS and MRS + cysteine agar plates using a subculture of bacterial suspension (10 μL per spot). The inoculated plates were incubated anaerobically at 37°C for 24 hours. Following incubation, MRS agar plates with Lactobacillus growth and MRS + cysteine agar plates with Bifidobacterium growth were overlaid with soft Mueller-Hinton agar (Biokar, France, BK048HA) pre-mixed with 100 μL (McFarland 1) of pathogenic strains. After solidification of the overlay medium, the plates were further incubated at 37°C for 24 hours. Pathogen inhibition was assessed by measuring the diameters of the inhibition zones (ZDI), which were interpreted according to Halder et al. (2017). Inhibition zones greater than 20 mm were classified as strong, those between 11–20 mm as intermediate, and those below 10 mm as weak. Additionally, agar well diffusion assays were performed to evaluate the antimicrobial activity of cell-free supernatants, following the method described by Fontana et al. (2013). The antibacterial activity of Lactobacillus and Bifidobacterium cell-free supernatants was tested against the previously mentioned multidrug-resistant bacteria.
- • Growth and Physiological Characterization of Bacterial Strains
To comprehensively evaluate the environmental adaptability of the isolated Lactobacillus and Bifidobacterium strains, a series of in vitro assays were conducted to assess their tolerance to varying temperatures, sodium chloride concentrations, and simulated gastrointestinal conditions. These tests aimed to determine the resilience of the strains under conditions mimicking those encountered in food processing, storage, and the human digestive tract. Temperature tolerance was assessed by incubating bacterial cultures at 15°C, 30°C, and 37°C in both MRS broth and MRS broth supplemented with 0.05% cysteine-HCl to support anaerobic bacterial growth. The rationale behind selecting these temperatures was to evaluate the adaptability of the strains under different environmental conditions. The 37°C incubation was used as a reference, as it corresponds to human body temperature and ensures optimal growth for probiotics. The 30°C condition was tested to simulate ambient temperatures where probiotic formulations might be stored, while 15°C was chosen to assess bacterial viability under refrigeration-like conditions. After 48 hours of incubation at 30°C and four days at 15°C, bacterial growth was evaluated by measuring optical density (OD600) using a spectrophotometer and observing colony formation on MRS agar plates (Arboleya et al., 2011; Riaz Rajoka et al., 2017).
The tolerance of isolated strains to sodium chloride (NaCl) was determined using the method described by Abhijit et al. (2012). The strains were cultivated in MRS broth containing 6.5% NaCl, a concentration that mimics high-salinity conditions such as those found in fermented food products. After 24 hours of incubation at 37°C, bacterial growth was assessed based on turbidity measurements using a spectrophotometer. The ability to survive in high-salt environments is a crucial characteristic for probiotic strains, as it ensures their stability during food processing and storage. The ability of bacterial strains to survive in conditions mimicking those of the human stomach was performed using the technique described by (Bahri et al., 2014). Simulated gastric juice was prepared by adding 3g of pepsin to 1L of 0.5% NaCl solution. The preparation was adjusted to pH=1.5. One milliliter of each bacterial culture (109 cells/ml; OD=620 nm between 0.5 and 0.7) was added to 9 ml of simulated gastric juice. Next day, 0.1 mL of the seeded gastric juice was taken after 0 and 2 h of reaction between the solutions respectively, and then plated onto MRS and MRS + Cysteine - HCl agar plates. The number of viable bacteria was determined after 24 to 48 h of incubation under anaerobic conditions. The experiment was repeated three times. The survival rate was calculated using the following equation:
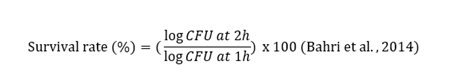
The ability of the strains to survive in conditions similar to those in the human small intestine was assessed using the technique described by (Bahri et al., 2014). For this, an intestinal juice was prepared as follows: 1 g/L pancreatin, was added to 0.5% NaCl, with or without 0.3% oxgall bile salts. Both preparations were adjusted to pH 8. Next, one milliliter of each bacterial culture (109 cells/ml) was inoculated into 9 mL of the two preparations. Finally, 0.1 ml of each preparation was taken at 0 h and 4 h of exposure and streaked onto MRS and MRS + Cysteine-HCl agar plates. Viable bacteria were counted after 24 to 48 h of incubation under anaerobic conditions. The experiment was repeated three times and the survival rate was calculated using the equation:
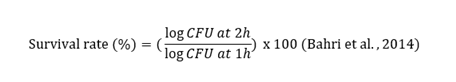
Data analysis
The results were expressed as mean ± standard deviation. Differences were considered significant at p < 0.05. Graphical presentations were generated using Excel and GraphPad Prism software version7
Results
Socio-demographic characteristic of studied population
The table 2 shown the socio-demographic data of studied population. From the 34 breastfeeding mothers enrolled in this study, the age group of [26 to 30 years old] represented 40% of the population, followed by the 21 to 25 age group, with a range of 10 to 50 years old. The average age was 27 years old. Among the women included in the study, 24.25% had experienced health problems such as hepatitis, an inflammation of the liver often caused by a viral infection, and ectopic pregnancies, where the embryo develops outside the uterus, usually in a fallopian tube. 15,15% have received antibiotic therapy for at least six months. Additionally, 45.45% had experienced signs of mastitis. Over 70% of women had no medical history. Moreover, over 80% had not undergone antibiotic therapy the last six months. Regarding the educational level of women, 76% have completed secondary education, and these women predominantly live in cohabitation.
Table 2: Socio-demographic data of population study
|
Parameters |
Percentage |
|
|
Breakdown of mothers by age |
||
|
[10-15] |
6 |
|
|
[16-20] |
9 |
|
|
[21-25] |
24 |
|
|
[26-30] |
40 |
|
|
[31-35] |
12 |
|
|
[36-40] |
3 |
|
|
[41-45] |
3 |
|
|
[46-50] |
3 |
|
|
Distribution of women by ethnic group |
||
|
Fang |
18 |
|
|
Mpongwe |
15 |
|
|
Punu |
43 |
|
|
Bakota |
3 |
|
|
Obamba |
3 |
|
|
Expatriate |
18 |
|
|
Level instruction |
||
|
Primary level |
6 |
|
|
Secondary |
76 |
|
|
Higher |
15 |
|
|
Illiterates |
3 |
|
|
Others parameters |
||
|
Medical history |
Yes |
24.25 |
|
No |
75.75 |
|
|
Antimicrobial treatment in the last 6 months |
Yes |
15.15 |
|
No |
84.85 |
|
|
Signs of mastitis |
Yes |
45.45 |
|
No |
54.55 |
|
Breastfeeding women's knowledge of breast milk composition and the benefits of breastfeeding was assessed, and the data are presented in Table 3. From this table, it emerges that women have a good understanding of breast milk composition and its benefits. It also shows that 66.66% of women are aware of the need for exclusive breastfeeding for at least 6 months, as recommended by the World Health Organization (WHO) while 33.33% have a poor understanding of this guideline.
Table 3: Women's knowledge of breastfeeding
|
Parameters |
Knowledgeable (%) |
Less Knowledgeable (%) |
|
Composition of breast milk |
50 |
50 |
|
Digestion of breast milk |
80 |
20 |
|
Antibodies in breast milk |
90 |
10 |
|
Allergies |
40 |
60 |
|
Protective role of breast milk |
65 |
35 |
|
Benefits of breast milk for babies |
90 |
10 |
|
Improvement of the immune system |
50 |
50 |
|
Duration of exclusive breastfeeding |
66.66 |
33.33 |
Isolation and biochemical and molecular identification of bacterial species from milk samples
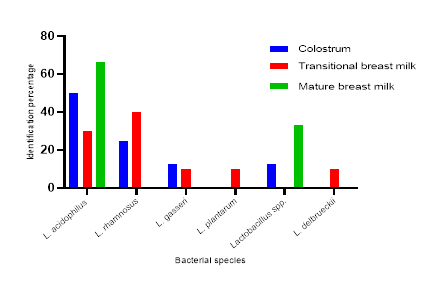
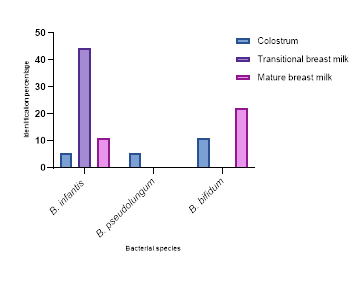
49 bacterial strains were isolated, of which 28 (57.14%) were Lactobacilli and 21 (42.86%) Bifidobacteria. Figures 1 and 2 present identification of bacteria according to the different types of milk, respectively for the genus Lactobacillus and the genus Bifidobacterium. Our data show the highly diversity of bacteria in colostrum. From Figure 1, it appears that colostrum and transitional milk have more bacteria than mature milk. Lactobacillus, L. acidophilus is the most representative species as it is found in all three types of milk in significant proportions: 30% in transitional milk, 50% in colostrum, and 66.7% in mature milk. From Figure 2, it is evident that among Bifidobacteria, B. infantis is present in all types of milk in highly variable proportions: 44.5% in transitional milk, 5.5% in colostrum, and 11.1% in mature milk. Only three species of Bifidobacterium were isolated from all three types of milkincluding B. infantis, B. pseudolungum, and B. bifidum. Among the Lactobacilli, Lactobacillus acidophilus was the most representative species in colostrum, transitional breast milk and mature breast milk. As for the Bifidobacteria, only the species Bifidobacterium infantis was isolated from the three types of breast milk, while Bifidobacterium pseudolungum was only isolated from colostrum.
Antibiotic susceptibility testing of isolated bacteria
The sensitivity of Lactobacilli and Bifidobacteria strains to antibiotics was assessed using the disc diffusion method. The resistance or susceptibility of a strain was determined by measuring the inhibition zone around the discs. From our results it appears that the strains are highly resistant to the tested antibiotics. Lactic acid bacteria are therefore naturally resistant to many antibiotics. But some strains such as Lactobacillus spp, L. plantarum and L. gasseri are sensitive to Streptomycin, Gentamicin and Ampicillin (Table 4).
Table 4: Resistance of potential probiotic bacteria isolated to antibiotics.
|
Antibiotics Disc |
Charge |
Lactobacillus |
L. |
L. |
B. |
B. |
|
spp |
plantarum |
gasseri |
bifidum |
infantis |
||
|
Penicilin |
10U/L |
R |
R |
R |
R |
R |
|
Ampicillin |
10µg |
S |
S |
S |
R |
R |
|
Oxacillin |
1µg |
R |
R |
R |
R |
R |
|
Cefoxitin |
30µg |
R |
R |
R |
R |
R |
|
Vancomycin |
30µg |
R |
R |
R |
R |
R |
|
Streptomycin |
10µg |
S |
S |
S |
R |
R |
|
Gentamicin |
10µg |
S |
S |
S |
R |
R |
|
Erythromycin |
15µg |
R |
R |
R |
R |
R |
|
Tetracyclin |
30µg |
R |
R |
R |
R |
S |
|
Clindamycin |
2µg |
R |
R |
R |
R |
R |
|
Chloramphenicol |
30µg |
R |
R |
R |
R |
R |
|
Nalidixic Acid |
30µg |
R |
R |
R |
R |
R |
|
Trimetroprim |
5µg |
R |
R |
R |
R |
R |
Legend: R = Resistant, S = Sensitive
Evaluation of probiotic capacity in vitro
Antimicrobial activity
The antibacterial activity of Lactobacilli and Bifidobacteria strains was assessed using the agar well diffusion method against clinical isolates of Salmonella typhimurium, Staphylococcus aureus, and the reference strain Escherichia coli ATCC 25922. This evaluation aimed to test the antibacterial potential of bacteria isolated from breast milk with high probiotic potential. Specifically, the antibacterial activity of L. gasseri (26C), Lactobacillus spp. (27C), L. plantarum (21M), B. bifidum (21M), and B. infantis (2C) was tested against all indicator strains, including both Gram-positive and Gram-negative bacteria. Inhibition zones greater than 6 mm were considered indicative of a positive or strong antagonistic effect. The most potent antimicrobial activity was observed with the B. bifidum (21M) strain, which exhibited inhibition zones exceeding 10 mm against all tested pathogens (Table 5). Additionally, the antimicrobial properties of other breast milk-derived bacteria with lower probiotic potential were evaluated but demonstrated weaker activity (data not shown).3.4.2. Capacity of bacteria to grow in a range of temperatures.
Table 5: Antibacterial activity profile of strains with high probiotic potential and their cell-free supernatant (CFS) against pathogenic bacteria
|
Inhibition diameter (mm) |
|||
|
Bacteria Strains |
P1=Salmonella typhimurium |
P2=Staphylococcus aureus |
Escherichia coli |
|
ATCC 25922 |
|||
|
L. gasseri |
8 |
8 |
8 |
|
CFS L. gasseri |
0 |
6 |
0 |
|
Lactobacillus spp |
8 |
8 |
8 |
|
CFS Lactobacillus. spp |
0 |
6 |
0 |
|
L. plantarum |
8 |
10 |
8 |
|
CFS L. plantarum |
0 |
7 |
0 |
|
B. infantis |
10 |
10 |
10 |
|
CFS B. infantis |
0 |
6 |
0 |
|
B. bifidum |
11 |
12 |
10 |
|
CFS B. bifidum |
0 |
7 |
0 |
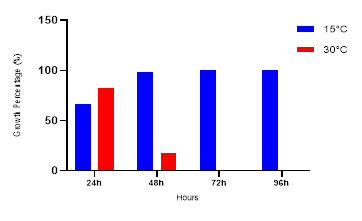
The isolated bacteria demonstrated the ability to grow across a wide range of temperatures (Figure 3). However, at 30°C, a decline in bacterial count was observed after 48 hours of incubation, although they remained viable at higher temperatures within the first 24 hours. In this study, all tested strains exhibited growth at both 15°C and 30°C for 48 hours, with a noticeable reduction at 30°C. These findings confirm that both Lactobacillus and Bifidobacterium genera are mesophilic in nature.
Growth Potential of Isolated Bacterial Strains.
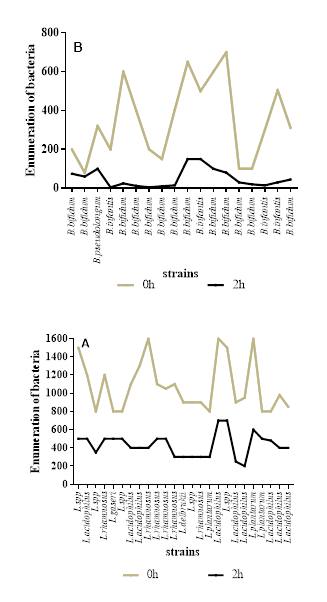
The figure 4 demonstrates that all the bacteria in the study have the potential to grow in a hostile environment. The strains isolated here were mostly resistant to NaCl at 6.5%.
Ability to survive in simulated intestinal conditions.
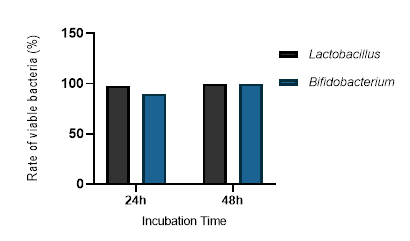
The bacterial strains exhibiting the highest resistance to simulated gastric juice were further evaluated for their ability to survive under simulated human small intestine conditions at pH 8, in the presence of pancreatin. After 4 hours of exposure, bacterial viability was assessed by counting colony-forming units on MRS and MRS + 0.05% cysteine agar plates. The results, presented in Figure 5, indicate a significant reduction in bacterial viability under simulated intestinal conditions. Particular attention was given to ten strains that demonstrated strong resistance to low pH, as our objective was to determine whether intestinal conditions had any impact on their survival and to confirm their resilience not only to acidic stress but also to other harsh conditions of the human intestine. Notably, after overcoming acidic conditions, these ten strains maintained or even enhanced their resistance when exposed to intestinal conditions, including elevated pH and pancreatin presence. Figure 5A illustrates the growth of Lactobacillus at pH 8, while Figure 5B presents the growth of Bifidobacterium under similar conditions.
Ability to survive in gastric conditions in the presence of bile salts
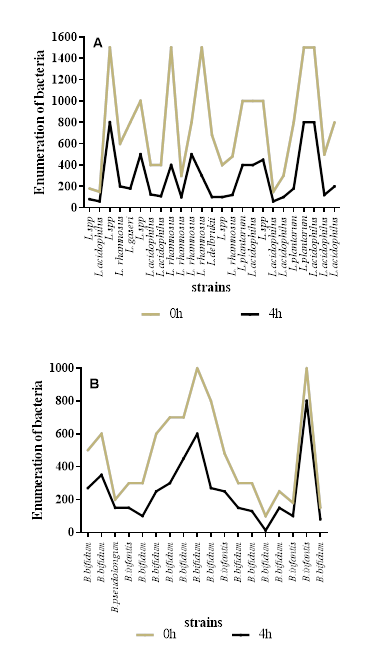
The bacterial strains were subjected to simulated gastric conditions (pH 1.5 with 3 g/L pepsin) to evaluate their survival. The viability of the isolated lactic acid bacteria was assessed after 1 and 2 hours of exposure, considering the combined effects of low pH and pepsin. We focused on ten strains that exhibited resistance at pH 1.5 to determine their ability to withstand these harsh gastric conditions compared to their survival at the same pH without pepsin. The selection of these ten strains was based on the premise that strains unable to survive at low pH alone would not endure similar conditions in the presence of pepsin. Under these conditions, all ten strains displayed resistance to both pepsin and acidic stress, although their viability decreased compared to exposure to low pH alone (Figure 6).
In summary, our study highlights that lactic acid bacteria (Lactobacillus and Bifidobacterium) isolated from the breast milk of Gabonese mothers exhibit significant resistance to acidic conditions and bile, underscoring their probiotic potential. They demonstrated strong inhibitory activity against S. aureus and E. coli in vitro, suggesting their potential health benefits. These findings emphasize the importance of breastfeeding in Gabon and the need to promote it as a protective measure against gastrointestinal diseases, particularly in low-income populations. Additionally, the antibacterial properties observed in this study contribute valuable insights into addressing antibiotic resistance.
Discussion
Lactic acid bacteria, which food habitat is milk and its derivatives, abound in a multitude of bacterial genera, including the Lactobacillus genus and the Bifidobacterium genus. Mother's milk is an essential and natural source of nutrition for infants, and its numerous health benefits addressing a range of infant health problems has long been recognized (Mehanna et al., 2013; Jamyuang et al., 2019). This study aimed to characterize the lactic acid bacteria (LAB) strains, specifically Lactobacillus and Bifidobacterium species, with probiotic potential in the breast milk of healthy breastfeeding women in Gabon. The results show that these strains exhibit promising probiotic properties, particularly Lactobacillus acidophilus and Bifidobacterium infantis, which are known for their resilience under gastrointestinal conditions.
Socio-demographic data showed that the level of knowledge about the importance of milk for both the infant and the mother was ascertained. From these data, it appears that the most of the breastfeeding women, aged 26 to 30 years old ,had no medical history has and possessed a secondary level of education. They were aware that breastfeeding contributes to the well-being of both mother and child. For example, these women knew that breastfeeding protects against certain diseases and aids in postpartum weight loss. They also acknowledged that breastfeeding plays a role in reducing the risk of postpartum hemorrhage and delaying the return of menstruation. We, therefore, believe that the promotion of breastfeeding through the description of breast milk components and its benefits will be a relevant strategy in public health. Also, mastitis, an inflammation of the breast tissue that affects over 45% of the women in this study, can influence the microbiota of breast milk. Inflammation in the breast tissue could alter the microbial composition and may have influenced the isolation of lactic acid bacteria, potentially biasing the results. Additionally, 20% of the women had undergone antibiotic therapy, which could further affect lactic acid bacteria isolation. Antibiotics may reduce the diversity of microbiota, including beneficial strains like Lactobacillus and Bifidobacterium, thereby skewing the findings. A future study should investigate the correlation between mastitis and lactic acid bacteria presence, as well as the effects of recent antibiotic use on lactic acid bacteria populations in breast milk.
Our data revealed that the lactic acid bacteria strains isolated from Gabonese breast milk possess promising probiotic properties. These bacteria, particularly Lactobacillus acidophilus and Bifidobacterium infantis, demonstrated the ability to withstand acidic conditions and bile, which is essential for their survival in the gastrointestinal tract and their effectiveness as probiotics. Lactobacillus acidophilus and Bifidobacterium infantis were the most representative species across different types of breast milk. This aligns with previous studies that indicate the prevalence of these bacterial species in breast milk and their potential role in maternal-infant health (Mehanna et al., 2013; Jamyuang et al., 2019). These results are also consistent with studies carried out in other countries such as Slovenia, the United States, Brazil and Algeria where more advanced techniques were used to isolate and characterize lactic acid bacteria belonging to the genera Lactobacillus and Bifidobacterium (Tušar et al., 2014; Rodrigues et al., 2020). Among the Lactobacillus species, Lactobacillus acidophilus was found to be the most predominant species in colostrum, transitional milk, and mature breast milk. This suggests that Lactobacillus acidophilus is a dominant species in breast milk, regardless of the lactation stage. In contrast, among the Bifidobacteria species, only Bifidobacterium infantis was isolated from all three types of breast milk, while Bifidobacterium pseudolungum was only found incolostrum. The presence of Lactobacillus acidophilus and Bifidobacterium infantis in breast milk is supported by previous research indicating the importance of these bacterial species in the colonization and development of the infant's intestinal microbiota (Riaz Rajoka et al., 2017; Blessing et al., 2020). The role of these probiotic bacteria in protection against necrotizing enterocolitis need further investigation(Lucas and Cole, 1990; Pammi et al., 2017; Langel et al., 2022). However, there are several methodological aspects that warrant further discussion and could influence the interpretation of the findings. One critical factor that was not addressed in this study is the potential impact of prior antibiotic treatments on the milk microbiota. Although the study participants were not undergoing any current medical treatments, antibiotics taken previously could still affect the microbial composition of breast milk. Unfortunately, the study did not define specific criteria for considering previous antibiotic treatments. Previous studies have suggested that antibiotics can significantly alter the composition of gut and milk microbiota, possibly diminishing the abundance of beneficial bacteria such as Lactobacillus and Bifidobacterium species. Therefore, a future study should include a more thorough assessment of participants' medical history and antibiotic use to control for this variable.
The isolated strains also exhibited significant antimicrobial activity against various pathogens, suggesting their therapeutic potential for improving infant gut health while their roles in gut-associated immunotolerance remain unexplored. Although some strains displayed antibiotic resistance, this did not hinder their antimicrobial efficacy, highlighting the need for further studies to understand the implications of this resistance in probiotic applications. However, this resistance could have implications for their safety and efficacy as probiotics. Although some strains displayed antibacterial activity against pathogens, the potential risks associated with the transfer of resistance genes, particularly in clinical settings, need to be carefully evaluated. Including a broader range of antibiotic classes in future analyses could provide more detailed insights into the resistance profiles of these strains and help guide their potential use in therapeutic applications. Lactobacillus acidophilus is known for its probiotic properties and its ability to support digestive health (Blessing et al., 2020). Its prevalence in breast milk suggests that it could contribute to establishing a healthy intestinal microbiota in breastfed infants, which is crucial for immune function and overall well-being. Similarly, Bifidobacterium infantis is recognized for its beneficial effects on infant health, including its role in promoting digestion, preventing infections, and modulating the immune system (Blessing et al., 2020). The consistent presence of Bifidobacterium infantis at different stages of lactation underscores its importance in early intestinal colonization and suggests that breast milk serves as a natural source of this beneficial bacterium for infants. Furthermore, the isolation of Bifidobacterium pseudolungum exclusively from colostrum highlights the dynamic nature of breast milk composition during the early stages of lactation, suggesting a particular role for this strain of bacteria (Blessing et al., 2020; Liu et al., 2020). Colostrum, also known as "first milk," is rich in bioactive components and is highly diversified. This milk provides essential nutrients and immune factors to newborns (Liu et al., 2020). The presence of specific bacterial species such as Bifidobacterium pseudolungum in colostrum may contribute to the early establishment of a diverse and beneficial gut microbiota in infants, thereby laying the groundwork for long-term health benefits (Blessing et al., 2020; Liu et al., 2020).
The study demonstrated that Lactobacilli and Bifidobacteria strains exhibit resilience to a wide range of environmental stressors, including temperature variations and high salt concentrations. Moreover, the strains displayed remarkable survival under simulated gastric and intestinal conditions, indicating their ability to withstand the hostile gastrointestinal environment. These findings underscore the potential of these bacterial strains as probiotics, capable of delivering health benefits to the host despite harsh physiological conditions in the digestive tract. The first requirement for a probiotic bacterium is its ability to survive transport to the active site in which its beneficial action is expected. Hence, the bacteria destined to benefit intestinal functions must survive passage through the acidic environment to the stomach (Pisano et al., 2014). In our study, their survival rate was higher than that of previously reported strains such as Lactobacillus rhamnosus and Lactobacillus casei ,under low pH conditions (Manini et al., 2016). And, antibiotic sensitivity assessment showed high resistance among Lactobacilli and Bifidobacteria strains, highlighting their intrinsic resistance to antibiotics. Moreover, despite antibiotic resistance, several strains exhibited significant antibacterial activity against pathogenic bacteria, indicating their potential as probiotics for therapeutic applications. Notably, Bifidobacterium bifidum strain exhibited the strongest antimicrobial activity, suggesting its promising role in combating bacterial infections especially gastrointestinal pathogens. The antibiotic resistance observed in some strains from our study raises important questions about their safe use as probiotics, particularly regarding the potential transfer of resistance genes. However, this resistance may also offer an advantage by allowing probiotics to persist in environments exposed to antibiotics, necessitating a thorough evaluation to balance the benefits and risks.
It would be beneficial to consider purifying these bacteria and exploring their therapeutic potential, particularly for preterm infants and those suffering from gastrointestinal diseases and diarrhea. However, our study has some limitations, including our inability to collect all three types of milk from each mother and the relatively small sample size. Additionally, a more comprehensive characterization of our strains using next-generation PCR will be recommended. Next-generation PCR can be used to amplify multiple genetic targets simultaneously, allowing for more efficient and precise identification of bacterial strains (Harris et al., 2010). This method could greatly improve the accuracy of strain identification and provide valuable data on the genetic diversity of LAB in breast milk. Incorporating this method into future studies would enhance the overall depth of the microbial analysis and lead to more reliable results.
In conclusion, while the study provides valuable insights into the probiotic potential of Lactobacillus and Bifidobacterium species in breast milk from Gabonese women, several methodological issues must be addressed in future research. These include considering the effects of prior antibiotic treatments, providing more detailed information on molecular identification and sequencing methods, and exploring the impact of mastitis and antibiotic use on lactic acid bacteria isolation. Furthermore, a broader range of environmental conditions, including temperature and antibiotic sensitivity, should be tested to better understand the potential of these lactic acid bacteria strains as probiotics. With these adjustments, future studies can better assess the therapeutic and preventive roles of these bacteria in infant health.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The authors declare no conflicts of interest.
Author Contributions
Conceptualization, G.L. and S.T.A; Methodology, N.M. ; Validation, G.L., S.T.A. and V.D.; Investigation, N.M. , K.K. , V.A., E.P.T. and P.E.N; Data curation, G.L., S.T.A., V.D. and L.B-M.; Writing—original draft preparation, B.B.L; Writing-review and editing, G.L., S.T.A., V.D. and L.B-M.; Supervision, , G.L., S.T.A., V.D. and L.B-M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding
Acknowledgments
We thank all the staff at the Maternal and Child Health Center of Port-Gentil (CSMI), as well as all the women who agreed to participate in the study.
Data Availability Statement
All data from this study are included in this manuscript.
References
- Abhijit C, Hossain M, Mostazir N, et al. Screening of Lactobacillus spp. from Buffalo Yoghurt for Probiotic and Antibacterial Activity. Bacteriol. Parasitol 3 (2012): 156.
- Agbankpe AJ, Dougnon TV, Balarabe R, et al. In vitro assessment of antibacterial activity from Lactobacillus spp. strains against virulent Salmonella species isolated from slaughter animals in Benin. Vet. World 12 (2019): 1951.
- Amadoro C, Rossi F, Pallotta ML, et al. Traditional dairy products can supply beneficial microorganisms able to survive in the gastrointestinal tract. LWT 93 (2018): 376–383.
- Arboleya S, Ruas-Madiedo P, Margolles A, et al. Characterization and in vitro properties of potentially probiotic Bifidobacterium strains isolated from breast-milk. Int. J. Food Microbiol. 149 (2011): 28–36.
- Arshad FA, Mehmood R, Hussain S, et al. Lactobacilli as Probiotics and their Isolation from Different Sources. Br. J. Res. 5 (2018).
- Bahri F, Lejeune A, Dubois-Dauphin R, et al. Characterization of Lactobacillus strains isolated from Algerian children faeces for their probiotic properties. Afr. J. Microbiol. Res. 8 (2014): 297–303.
- Blessing EN, Chukwuemeka I, Ukeachu CD, et al. Antibacterial properties of probiotics bacterial isolated from human breast milk.
- Recommandations 2022.
- Chong HY, Tan LTH, Law JWF, et al. Exploring the potential of human milk and formula milk on infants’ gut and health. Nutrients 14 (2022): 3554.
- D’Alessandro M, Parolin C, Patrignani S, et al. Human Breast Milk: A Source of Potential Probiotic Candidates. Microorganisms 10 (2022): 1279.
- Essomo MMMEO, Abessolo JIO, Abaga FM, et al. Composition du lait maternel chez la femme gabonaise au cours des trois premieres semaines d’allaitement. Health Sci. Dis. (2013): 1–5.
- Fontana L, Bermudez-Brito M, Plaza-Diaz J, et al. Sources, isolation, characterisation and evaluation of probiotics. Br. J. Nutr. 109 (2013): S35–S50.
- Fortmann I, Marißen J, Siller B, et al. Lactobacillus acidophilus/bifidobacterium infantis probiotics are beneficial to extremely low gestational age infants fed human milk. Nutrients 12 (2020): 850.
- Gunyakti A, Asan-Ozusaglam M. Lactobacillus gasseri from human milk with probiotic potential and some technological properties. LWT 109 (2019): 261–269.
- Hadadji M. Caractérisation Microbiologique et Biochimique des Bifidobactéries isolées à partir des selles de nourrissons. Faculté des Sciences: Université d’Oran, Es-Sénia (2007).
- Jamyuang C, Phoonlapdacha P, Chongviriyaphan N, et al. Characterization and probiotic properties of Lactobacilli from human breast milk. 3 Biotech 9 (2019): 398.
- Javed GA, Arshad N, Munir A, et al. Signature probiotic and pharmacological attributes of lactic acid bacteria isolated from human breast milk. Int. Dairy J. 127 (2021): 105297.
- Juharji H, Albalawi K, Aldwaighri M, et al. Impact of Breastfeeding on Low Birthweight Infants, Weight Disorders in Infants, and Child Development. Cureus 14 (2022): e32894.
- Langel SN, Blasi M, Permar SR. Maternal immune protection against infectious diseases. Cell Host Microbe 30 (2022): 660–674.
- Liu W, Chen M, Duo L, et al. Characterization of potentially probiotic lactic acid bacteria and bifidobacteria isolated from human colostrum. J. Dairy Sci. 103 (2020).
- Lokossou GAG, Kouakanou L, Schumacher A, et al. Human Breast Milk: From Food to Active Immune Response With Disease Protection in Infants and Mothers. Front. Immunol. 13 (2022).
- Łubiech K, Twarużek M. Lactobacillus bacteria in breast milk. Nutrients 12 (2020): 3783.
- Lucas A, Cole TJ. Breast milk and neonatal necrotising enterocolitis. Lancet 336 (1990): 1519–1523.
- Lyons KE, Ryan CA, Dempsey EM, et al. Breast Milk, a Source of Beneficial Microbes and Associated Benefits for Infant Health. Nutrients 12 (2020): 1039.
- Manini F, Casiraghi MC, Poutanen K, et al. Characterization of lactic acid bacteria isolated from wheat bran sourdough. LWT - Food Sci. Technol. 66 (2016): 275–283.
- Mehanna NSH, Tawfik NF, Salem MME, et al. Assessment of Potential Probiotic Bacteria Isolated from Breast Milk (2013).
- Mohammadi F, Eshaghi M, Razavi S, et al. Characterization of bacteriocin production in Lactobacillus spp. isolated from mother’s milk. Microb. Pathog. 118 (2018): 242–246.
- Moossavi S, Azad MB. Origins of human milk microbiota: new evidence and arising questions. Gut Microbes 12 (2020): 1667722.
- Nikolopoulou G, Tsironi T, Halvatsiotis P, et al. Analysis of the Major Probiotics in Healthy Women’s Breast Milk by Realtime PCR. Appl. Sci. 11 (2021): 9400.
- Oddi S, Huber P, Duque ARF, et al. Breast-milk derived potential probiotics as strategy for the management of childhood obesity. Food Res. Int. 137 (2020): 109673.
- Pammi M, Cope J, Tarr PI, et al. Intestinal dysbiosis in preterm infants preceding necrotizing enterocolitis: a systematic review and meta-analysis. Microbiome 5 (2017): 31.
- Park J, Yi DY. Comprehensive analysis of the effect of probiotic intake by the mother on human breast milk and infant fecal microbiota. J. Korean Med. Sci. 36 (2021).
- Pisano MB, Viale S, Conti S, et al. Preliminary evaluation of probiotic properties of Lactobacillus strains isolated from Sardinian dairy products. BioMed Res. Int. 2014 (2014): 286390.
- Riaz Rajoka MS, Mehwish HM, Siddiq M, et al. Identification, characterization, and probiotic potential of Lactobacillus rhamnosus isolated from human milk. LWT 84 (2017): 271–280.
- Rodrigues JZS, Passos MR, Silva de Macêdo Neres N, et al. Antimicrobial activity of Lactobacillus fermentum TcUESC01 against Streptococcus mutans UA159. Microb. Pathog. 142 (2020): 104063.
- Rodríguez Arreola A, Solís Pacheco JR, Lacroix M, et al. In vivo assessment and characterization of lactic acid bacteria with probiotic profile isolated from human milk powder. Nutr. Hosp. 38 (2020): 152–160.
- Sakwinska O, Moine D, Delley M, et al. Microbiota in Breast Milk of Chinese Lactating Mothers. PLOS ONE 11 (2016): e0160856.
- Selvamani S, Dailin DJ, Gupta VK, et al. An Insight into Probiotics Bio-Route: Translocation from the Mother’s Gut to the Mammary Gland. Appl. Sci. 11 (2021).
- Shokryazdan P, Faseleh Jahromi M, Liang JB, et al. Probiotics: From Isolation to Application. J. Am. Coll. Nutr. 36 (2017): 666–676.
- Tušar T, Žerdoner K, Matijašić BB, et al. Cultivable Bacteria from Milk from Slovenian Breastfeeding Mothers. Food Technol. Biotechnol. (2014).
- Zacarías MF, Binetti A, Laco M, et al. Preliminary technological and potential probiotic characterisation of bifidobacteria isolated from breast milk for use in dairy products. Int. Dairy J. 21 (2011): 548–555.
- Zuo F, Yu R, Feng X, et al. Characterization and in vitro properties of potential probiotic Bifidobacterium strains isolated from breast-fed infant feces. Ann. Microbiol. 66 (2016): 1027–1037.


 Impact Factor: * 3.5
Impact Factor: * 3.5 Acceptance Rate: 71.36%
Acceptance Rate: 71.36%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 
