Phenotypic and Genotypic Virulence Factors and Their Association with Antibiotic Resistance in Clinical Isolates of Enterococcus Species in Bangladesh
Tahani Momotaz1, 2, Rehana Razzak Khan1, Fatima Afroz1, Sharmin Chowdhury1, Md. Nahidul Islam2, Mohammad Tanvir Sarwar*3, 4, Abu Naser Ibne Sattar1
1Department of Microbiology and Immunology, Bangabandhu Sheikh Mujib Medical University, Shahbag, Dhaka-1000, Bangladesh
2Department of Microbiology, Kushtia Medical College, Kushtia, 7000 Bangladesh
3Department of Applied Nutrition and Food Technology, Faculty of Biological Sciences, Islamic University Kushtia, 7003 Bangladesh
4Department of Oral Biology, School of Dental Medicine, State University of New York at Buffalo, Buffalo, New York 14214-8024, USA
*Corresponding author: Mohammad Tanvir Sarwar, Department of Oral Biology, School of Dental Medicine, State University of New York at Buffalo, Buffalo, New York 14214-8024, USA.
Received: 27 July 2024; Accepted: 31 July 2024; Published: 06 August 2024
Article Information
Citation:
Tahani Momotaz, Rehana Razzak Khan, Fatima Afroz, Sharmin Chowdhury, Md. Nahidul Islam, Mohammad Tanvir Sarwar, Abu Naser Ibne Sattar. Phenotypic and Genotypic Virulence Factors and Their Association with Antibiotic Resistance in Clinical Isolates of Enterococcus Species in Bangladesh. Archives of Microbiology and Immunology. 8 (2024): 376-382.
View / Download Pdf Share at FacebookAbstract
Introduction: The emergence of drug resistant Enterococcus spp is now become an important public health threat as it is one of the leading causes of nosocomial infections. This study aimed to disclose the virulence factors and their encoding genes (asa, gelE, esp, ebpR, hyl gene for biofilm; cylA gene for hemolysis; gelE gene for gelatin hydrolysis) and observe their relations with antimicrobial resistance in Enterococci.
Methods: For this cross-sectional study, a total of 87 Enterococci isolated from different clinical samples (urine, blood, wound swab, pus and bile) were collected. Virulence factors were detected phenotypically by observing hemolysis, gelatin hydrolysis and biofilm formation by tissue culture plate method. For detection of virulence genes, conventional multiplex PCR was adopted for all genes except ebpR gene which was identified by single conventional PCR. Kirby Bauer disc diffusion method was used for antimicrobial susceptibility testing.
Results: Among the isolated Enterococci majority were E. faecalis (75%) followed by E. faecium (23%) and E. raffinosus (2%). About 52.3% of E. faecalis and 35% of E. faecium isolates were biofilm producers. Both in E. faecalis and in E. faecium significant association were found between biofilm formation and asa, ebpR, esp genes (p value<.05). Hemolysis was phenotypically observed in 30.8% isolates of E. faecalis and 20% isolates of E. faecium. A significant association was observed between cylA gene and hemolysin production in E. faecalis. Eighty five percent, 52.9% and 70.6% biofilm producing and 80%, 55% and 80% hemolysin producing isolates of E. faecalis were resistant to ciprofloxacin, ampicillin and gentamicin respectively which was statistically significant (p value <.05).
Conclusion: Antibiotic resistance was higher in biofilm and hemolysin producing isolates of both species. All biofilm producing isolates of E. faecalis and E. faecium were sensitive to vancomycin, linezolid, teicoplanin and fosfomycin.
Keywords
<p>Enterococci, biofilm, hemolysin, gelatinase, virulence genes, PCR</p>
Article Details
1. Introduction
Enterococci are common residents of the gastrointestinal tracts of humans and animals which causes various community and hospital acquired infections such as urinary tract infections, bacteremia, endocarditis, meningitis and intra-abdominal infections [1]. Enterococci are considered as important nosocomial pathogens because they have the capacity to acquire and share extra chromosomal elements including antibiotic resistance genes or virulence traits. Virulence factors of Enterococci might have a role in increasing their capacity to colonize among hospitalized patients [2]. The ability of biofilm formation which protect the bacteria from host immune responses and antibiotics is one of the important virulent characteristics of Enterococci [3]. Numerous enterococcal virulence factors including secreted factors and adhesions such as Esp (extracellular surface protein), Asa 1 (aggregation substance) and Ebp (endocarditis and biofilm-associated pili) are related with the biofilm formation [4]. Asa1, a pheromone-inducible protein escalates bacterial attachment to renal tubular cells and Esp (cell wall-associated protein) helps in colonization, persistence and biofilms formation in the urinary system [5]. The expression of Ebp operon is regulated by ebpR (endocarditis and biofilm associated pilli regulator) gene and associated with pilli and biofilm formation in enterococcal species causing UTI [6, 7]. Biofilm formation is reduced in ebp mutant strains [6]. Several secreted virulence factors of Enterococci including CylA (cytolysin), GelEA (gelatinase) and Hyl (hyaluronidase) play a role in pathogenesis of Enterococci. Gelatinase (extracellular zinc-containing metalloproteinase) degrades host tissue, provides nutrients to the bacteria and helps in biofilm formation. Hyl acts on hyaluronic acid and facilitates the spread of Enterococci by damaging the host tissue with the help of its degradative enzyme [2]. Cytolysin (secreted toxin) is produced in response to pheromones and helps in the pathogenesis of E. faecalis by creating blood hemolysis [8]. Among Enterococcus species, E. faecalis is the most virulent and pathogenic strain which contains several effective mechanisms like highly transmissible plasmid for horizontal gene transfer that helps them to transmit virulence genes to other less virulent species as E. faecium [9]. Understanding the virulence factors of Enterococci may help to know the pathogenic process and antimicrobial resistance of this bacteria. So, this study profiled Enterococci isolates of clinical origin for their virulence factors, virulence encoding genes and their association with antibiotic resistance.
2. Materials and Methods
This cross-sectional study was conducted at the Department of Microbiology and Immunology, Bangabandhu Sheikh Mujib Medical University (BSMMU) over a period of one year from March 2019 to February 2020.
2.1 Bacteria isolates collection and identification
A total of 87 Enterococci isolated from different clinical samples (urine, blood, wound swab, pus and bile) were collected from the Laboratory of Microbiology and Immunology Department of BSMMU for this study. These samples were sent from different departments of BSMMU hospital for culture and sensitivity test. Enterococci were identified based on colony morphology on Chromogenic agar media and blood agar media, Gram staining method and standard biochemical test (catalase test, salt tolerance test, bile esculin test) [10]. Then identification of Enterococci species was done by fermentation of mannitol, sorbitol, raffinose, arabinose, utilization of pyruvate and arginine decarboxylase test [11].
2.2 Antibiotic susceptibility test by disc diffusion
For antimicrobial susceptibility testing of the isolated Enterococci, the Kirby-Bauer disk diffusion method was used using Mueller-Hinton agar (Himedia, India) and commercially available antibiotic discs (Biomaxima, Poland). Antibiotic discs of ampicillin (10 µg), ciprofloxacin (5 µg), cotrimoxazole (1.25/23.75 µg), nitrofurantoin (300 µg), gentamicin (120 µg), vancomycin (30 µg), linezolid (30 µg), teicoplanin (30 µg), fosfomycin (200 µg), and quinopristin- dalfopristin (15 µg) were used. The disc concentration and zone of inhibition were used as recommended by the Clinical Laboratory Standards Institute guideline (CLSI, 2019) [12]. S. aureus ATCC 25923 was used as a quality control strain.
2.3 Phenotypic identification of the virulence factors
2.3.1 Detection of hemolysis
Enterococci were inoculated on blood agar media to detect the hemolytic activity and observed the appearance of hemolytic zone around the colony after 24 hours of incubation at 37°C [13].
2.3.2 Gelatin hydrolysis activity
Production of gelatin was detected by inoculation of Enterococci on freshly prepared peptone yeast extract agar containing 4% gelatin and incubated at 37°C for 24 hours. Then cooled to room temperature for two hours. Gelatin hydrolysis was indicated by the appearance of turbid halo around the colonies [14].
2.3.3 Biofilm formation
Biofilm formation of Enterococci was performed by tissue culture plate method (TCP) as previously described by Toledo-Arana et al [15]. At first, the Enterococci were grown in Brain heart infusion broth (BHIB) (Becton Dickinson and company, USA) with 0.25% glucose which was incubated at 37°C overnight. Then the broth culture was diluted at a ratio of 1:40. Two hundred microliter of diluted culture suspension was inoculated in a sterile 96 well flat bottom polystyrene micro titer plate (Greiner Bio-One International, Kremsmunster, Austria). The positive control (Klebsiella pneunoniae ATCC 700603) and negative control (sterile BHIB-0.25% glucose) were also inoculated in the same way. Then the wells were washed three times with 200 μl of phosphate buffer saline (PBS) after overnight incubation at 37°C. The plate was air dried, fixed with 200 μl/well of 2% formalin at 4ºC for 1 hour. The plate was stained with 1% crystal violet for 15 min and to remove the excess stain plate was rinsed under running tap water. After that 200 μl ethanol acetone (80:20, v/v) was added in each well to solubilize crystal violet. Each assay was performed in triplicate and repeated three times. The optical density (OD) at 630 nm was measured using ELISA plate reader (Plate reader, model–A4, serial no.-1910, Das, Italy). The cut-off value (ODc) was calculated for each microtiter plate. ODc was of three standard deviations (SD) above the mean OD of the negative control: ODc = average OD of negative control + (3×SD of negative control). Final OD value of a tested strain was expressed as average OD value of the strain reduced by ODc value (OD = average OD of a strain - ODc). Negative value presented as zero, while any positive value indicates biofilm production [16].
2.4 Genotypic detection of virulence genes by PCR
Extraction of DNA was performed by boiling method [17]. Conventional multiplex PCR was used to detect all virulence genes and single PCR was used to detect the ebpR gene using primer sets listed in Table 1.

The PCR assay was done in a reaction mixture with total volume of 25 μl which contained 15 μl of master mix (HELINI, India), 0.15 μl Taq polymerase (Solis BioDyne Germany), 1 μl of forward and reverse primer each (10 pmol/μl), 2.85 μl of distilled water and 5 μl of undiluted extracted DNA. For all virulence genes the amplification condition was: pre-denaturation at 95 °C for 15 min followed by 30 cycles of denaturation at 94 °C for 1 min, annealing at 56 °C for 1 min and extension at 72 °C for 1 min and final extension at 72 °C for 10 min. In case of ebpR the amplification condition was: pre- denaturation at 95 °C for 5 min followed by 35 cycles of denaturation at 95 °C for 1 min, annealing at 52 °C for 30 sec and extension at 72 °C for 50 sec and final extension at 72 °C for 10 min. The amplification products were electrophoresed on 1.5% agarose gel.
2.5 Statistical analysis
The qualitative data were expressed as frequency and percentages. The association between the biofilm formation, hemolysin production and their encoding genes with antibiotics resistance were assessed by the Pearson Chi-Square test using SPSS version 22. p values less than 0.05 were considered as statistically significant.
3. Results
Among the 87 Enterococci isolates, 75% (65) were E. faecalis, 23% (20) E. faecium and 2 % (2) E. raffinosus. Approximately 47% (41) Enterococci isolates were biofilm producers and 27.6 % (24) showed hemolysis. Among E. faecalis and E. faecium 52.3% (34) and 35% (7) of isolates were biofilm producers respectively. Twenty percent (4) and 30.8% (20) of isolates of E. faecium and E. faecalis showed hemolysis production respectively. Gelatin hydrolysis was not detected in any isolates [Table-2].

The association of single and combination of virulence factors encoding genes with phenotypic virulence factors in E. faecalis and E. faecium were shown in Table 3,4,5,6. The asa, esp and ebpR were found in 65.9%, 64.1% and 60.4% of biofilm producing E. faecalis (p value <.05). The combination of genes asa, gelE, ebpR and asa, esp, gelE, ebpR were also found in 87.5% & 86.4% of biofilm producing E. faecalis which were statistically significant [Table-3].

Hundred percent of asa, gelE, combination of esp, hyl, ebpR genes; 85.7% of esp gene and 50 % of ebpR gene were found in biofilm producing E. faecium isolates which were statistically significant (p value <.05) [Table-4].

Among 46 cylA positive E. fecalis isolates, 43.6% (20) were phenotypically hemolysin producers and all (19) cylA negative isolates were negative for hemolysis phenotypically which was statistically significant (p value 0.001) [Table-5]. The association of cylA gene and hemolysin production in E. faecium isolates were showed in Table 6. None of the hemolysin producing E. faecium isolate was positive for cylA gene. On the other hand, out of 20 cylA gene negative isolates, 20% (4) were hemolysin producers [Table-6].

Statistical analysis stipulated that there was a notable relationship between biofilm formation and antibiotic resistance in E. faecalis. Fifty three percent, 64.7%, 85.3% and 70.6% of biofilm producing E. faecalis isolates were resistant to ampicillin, cotrimoxazole, ciprofloxacin and gentamicin respectively (p< .05). Antibiotic resistance was higher in biofilm producing E. faecium but no significant association was found. All biofilm producing isolates of E. faecalis and E. faecium were sensitive to vancomycin, linezolid, teicoplanin and fosfomycin [Table-7, Table-8].


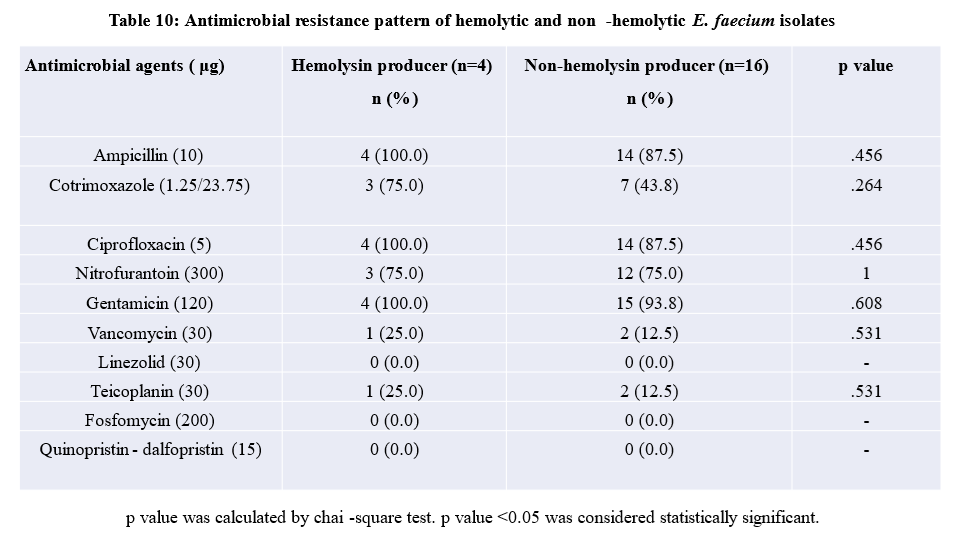
Eighty percent of hemolytic E. faecalis isolates were resistant to ciprofloxacin and gentamicin. Significant association was found in case of ampicillin, ciprofloxacin, nitrofurantoin and gentamicin resistance and hemolysin production in E. faecalis (p< .05). Hundred percent of hemolysin producing isolates of E. faecium were resistant to ampicillin, ciprofloxacin, gentamicin and 75% isolates were resistant to cotrimoxazole and nitrofurantoin but this association was not statistically significant [Table-9 and Table-10

Discussion
The increased incidence of drug resistant enterococcal infections in recent years suggests that several virulence factors might have a role in accelerating the pathogenesis and antibiotic resistance of this bacteria. So, it is very important to detect the virulence factors with their encoding genes and their association with antimicrobial resistance in clinical setting for epidemiological surveillance, formulation of local antibiogram, treatment guideline, infection control and preventive measures. Among 87 Enterococci isolates, 75% were E. faecalis followed E. faecium (23%) and E. raffinosus (2%). Biofilm-producing Enterococci are responsible for recurrent, chronic and antibiotic-resistant infection [19]. In biofilm producing bacteria the matrix component of biofilm reduces the penetration of antibiotic, presence of persister cells and the horizontal gene transfer of antibiotic resistance genes leads to antibiotic resistance [20, 21]. Approximately 47.1% of Enterococci isolates were biofilm producers which is almost similar to the findings (32.4%) reported in a study of Eastern India [22]. On the contrary in another study, lower percentage (21.9%) of biofilm producing isolates were reported where less virulent E. faecium was the predominant isolates [23].
In our study, 52.3% of E. faecalis and 35.0% of E. faecium isolates were biofilm producers. Similarly in a study of China, 47.2% of biofilm producing E. faecalis isolates were observed [24]. Some study reported that E. faecium produces more biofilms than E. faecalis [5]. The conflict may be due to type of sample, number of E. faecium isolates and geographic dissimilarities. Enterococci harbors plasmid pAD1 which encodes cytolysin and helps in the transfer of virulence factors and antibiotic resistance genes [8, 25]. In this study about 27.6% of hemolysin producing isolates of Enterococci were observed. Similarly, 34.2% of hemolysin producing Enterococci were reported by other studies [22]. Fifty eight percent of hemolysin producing Enterococci was reported in a study of West Iran which may be due to regional difference [26]. The role of gelatinase in enterococcal infection is to formation of biofilm and supply of nutrients to the bacteria [2]. None of the Enterococcus species was positive for gelatin hydrolysis and similar findings were also reported by studies done in Egypt. Many gelE positive isolates failed to secrete gelatinase which may be the cause of this findings [27]. Transmembrane protein, FsrB regulates the production of gelatinase by gelE gene which is controlled by locus fsr and deletions within locus fsr produce mutants that do not synthesize gelatinase [28]. Approximately 20% of gelatinase producing Enterococci isolates were reported in a study of Kolkata which may be due to infection with different strains of Enterococcus in that region [29].
In this study, 100.0 % of biofilm producing E. faecium and 65.9% of biofilm producing E. faecalis isolates had asa gene. Abdel et al also found the association of asa gene with biofilm formation but opposite findings were reported by Fallah et al [30, 5]. The esp gene was observed in 64.1% and 85.7% of biofilm producing E. faecalis and E. faecium respectively which was statistically significant (p value < .05). A correlation between the esp gene and biofilm formation was also observed by studies done in Saudi Arabia [30]. Some other studies suggest that the esp gene is not mandatory for the biofilm formation in E. faecium and E. faecalis [31]. Hundred percent and 53.8% of biofilm forming E. faecium and E. faecalis isolates had gelE gene where the association was statistically significant for E. faecium (p value < .05). Association of gelE gene with production of biofilm in Enterococci specially in E. faecium were reported by some studies but other study suggests that gelatinase was not necessary for biofilm production [32, 33].
Fifty percent and 60.4% of biofilm producing E. faecium and E. faecalis isolates had ebpR gene which was statistically significant (p <.05). Higher percentages (93.6%) of biofilm producing Enterococcus having ebp gene is also reported [4].
All biofilm producing E. faecium and E. faecalis isolates of this study had multiple biofilms forming genes. Combination of asa, gelE, ebpR genes and asa, esp, gelE, ebpR genes had significant relationship with biofilm formation (p <.05) in E. faecalis. We also found statistically significant (p < .05) association of combination of esp, ebpR and esp, hyl, ebpR genes with biofilm formation in E. faecium. This was supported by other studies where they found that single biofilm associated gene (esp, asa, gelE, ebpR) was not enough for biofilm development in Enterococci [5, 34]. On the contrary, other study reported that single biofilm forming gene was related with biofilm production [32]. Production of biofilm is a complicated process and relies on multiple factors in Enterococcus strains which may be the reason of different findings of the biofilm encoding genes with biofilm formation [35].
Cytolysin which is responsible for hemolytic activity has a key role in the extremity of human infections [36]. Hemolytic activity was observed phenotypically in 30.8% isolates of E. faecalis whereas cylA was present in 43.5% isolates which was statistically significant (p value 0.001). Similar findings were also reported in another study where they showed 41% of Enterococci carried the cylA gene and hemolytic activity was observed in 38% of cylA positive isolates [34]. This lack of phenotypic/genotypic expression of cytolysin might propose the mislay genes in the cyl operon [34]. The cylA gene was absent in all (4) hemolysin producing isolates of E. faecium. The cytolysin structural gene was always detected in β-hemolytic E. faecalis while absent in E. faecium [37]. So, the hemolysis of E. faecium must be occured by another cytotoxic component. In a study of South Korea, cylA gene negative E. faecium isolates with hemolytic activity were detected which suggests possible function of other genes in hemolytic activity [38].
Treatment of biofilm forming Enterococci are difficult because they are more antibiotic resistant [24]. Regarding antibiotic resistance in E. faecalis, higher resistance was observed in biofilm producers compared to non-biofilm producers. Resistance to some antibiotics including ampicillin (52.9% vs. 12.9%), cotrimoxazole (64.7% vs. 16.1%), ciprofloxacin (85.3% vs. 35.5%), gentamicin (70.6% vs. 29.0%) were significantly higher in biofilm producing E. faecalis than non-biofilm producers. Similar results were reported by Fallah et al and they found significantly higher resistance among biofilm positive isolates [5]. Biofilm producing E. faecium isolates were 100.0% resistant to ampicillin, ciprofloxacin, gentamicin followed by 71.4% resistant to cotrimoxazole and nitrofurantoin. Biofilm producing E. faecium strains were more antibiotic resistant [39]. Hemolysin producing E. faecalis isolates of this study were more antibiotic resistant than non-hemolysin producing isolates. Resistance to some antibiotics including ampicillin (55% vs. 24.4%), ciprofloxacin (80% vs. 53.3%), gentamicin (80% vs. 37.8%), nitrofurantoin (30% vs. 4.4%) were significantly higher in hemolysin producer than non-hemolysin producers.
Resistance to various antibiotics including ampicillin (100% vs. 87.5%), cotrimoxazole (75% vs. 43.8%), ciprofloxacin (100% vs. 87.5%), gentamicin (100% vs. 93.8%) were higher in hemolysin producer than non-hemolysin producers of E. faecium. Hemolysin producing isolates were more antibiotic resistant than non-hemolysin producing isolates but no significant association was observed and similar findings were also reported by Jankoska et al [40]. Higher number of resistances were also seen in non-hemolysin and non-biofilm producing isolates of E. faecium. The reason may be due to E. faecium is more drug resistant.
Conclusion
This study reveals the ability of Enterococci to develop biofilm formation and hemolysin production. The biofilm encoding genes ebpR, asa, esp, gelE were found in biofilm producing isolates of both species and hemolysin encoding gene cylA in hemolysin producing E. faecalis isolates which suggests the potential link between these genes in biofilm formation and hemolysin production. Vancomycin, linezolid, fosfomycin and teicoplanin remains the most effective antimicrobial agents in hemolysin and biofilm producing isolates of E. faecalis and E. faecium.
Acknowledgement:
The corresponding author is grateful to the Department of Microbiology and Immunology, Bangabandhu Sheikh Mujib Medical University (BSMMU) for arranging necessary facilities to perform the research.
Recommendations
Further nationalwide large scale study should be explored to know the prevalence of Enterococcal infection in Bangladesh.
Ethical Approval:
The study was ethically approved by Institutional Review Board (IRB) of Bangabandhu Sheikh Mujib Medical University, Dhaka, Bangladesh [NO.BSMMU/2019/8188, Date- 29/07/2019]
Conflict of Interest:
The authors declare that they have no conflicts of interest.
Author’s Contribution
Abu Naser Ibne Sattar was responsible for the conception of the study. Abu Naser Ibne Sattar, Rehana Razzak Khan, Fatima Afroz, Md. Nahidul Islam, Sharmin Chowdhury, Tahani Momotaz and Mohammad Tanvir Sarwar participated in its design and coordination. Tahani Momotaz was chief investigator and responsible for the acquisition, analysis and interpretation of the data. Mohammad Tanvir Sarwar, Abu Naser Ibne Sattar, Fatima Afroz and Rehana Razzak Khan reviewed the results and statistical analyses. Tahani Momotaz and Fatima Afroz drafted the manuscript and all the authors contributed substantially to its revision. All authors met ICMJE authorship criteria and have read and approved the final manuscript.
References
- Bourdon N, Fines-Guyon M, Thiolet JM, Maugat S, Coignard B, Leclercq R, et al. Changing trends in vancomycin-resistant enterococci in French hospitals, 2001–08. J Antimicrob Chemother 66 (2011): 713–721.
- Suchi SE, Shamsuzzaman SM, Uddin BM, Yusuf MA. Detection of virulence factors and antimicrobial resistance in enterococci isolated from urinary tract infection. Bangladesh Journal of Infectious Diseases 4 (2017): 30.
- Guzman-Soto I, McTiernan C, Gonzalez-Gomez M, Ross A, Gupta K, Suuronen EJ, et al. Mimicking biofilm formation and development: Recent progress in in vitro and in vivo biofilm models. iScience 24 (2021): 102443.
- Kafil HS, Mobarez AM. Assessment of biofilm formation by enterococci isolates from urinary tract infections with different virulence profiles. King Saud Univ.-Sci 27 (2015): 312-7.
- Fallah F, Yousefi M, Pourmand MR, Hashemi A, Alam AN and Afshar D. Phenotypic and genotypic study of biofilm formation in Enterococci isolated from urinary tract infections. Microbial pathogenesis 108 (2017): 85-90.
- Bourgogne A, Singh KV, Fox KA, Pflughoeft KJ, Murray BE and Garsin DA. EbpR is important for biofilm formation by activating expression of the endocarditis and biofilm-associated pilus operon (ebpABC) of Enterococcus faecalis Journal of bacteriology 189 (2007): 6490-93.
- Kafil HS, Mobarez AM, Moghadam MF. Adhesion and virulence factor properties of Enterococci isolated from clinical samples in Iran. Indian J. Pathol. Microbiol 56 (2013): 238-42.
- Ike Y, Hashimoto H, Clewell DB. Hemolysin of Streptococcus faecalis subspecies zymogenes contribute to virulence in mice. Infect Immun 45 (1984): 528–530.
- Biswas PP, Dey S, Sen A, Adhikari L. Molecular Characterization of Virulence Genes in Vancomycin-Resistant and Vancomycin-Sensitive enterococci. J Glob Infect Dis 8 (2016): 16-24.
- Boby F, Ahmed S, Paul S, Nasreen S, Haque N, Roy S. Antimicrobial susceptibility pattern of Enterococci isolated from clinical specimens at Mymensingh Medical College Hospital, Mymensingh. J Bacteriol Mycol Open Access 3 (2016): 241-4.
- Facklam RR, Collins MD. Identification of Enterococcus species isolated from human infections by a conventional test scheme. J Clin Microbiol 27 (1989): 731-4.
- Wayne P. Performance standards for antimicrobial susceptibility testing; 29th informational supplement. CLSI document M100-S29.
- Manavalan J, Kannaiyan K, Velayutham A, Vadivel S and Kuthalaramalingam S. Phenotypic speciation of Enterococci with special reference to prevalence, virulence and antimicrobial resistance. Int J Res Med Sci 3 (2015): 2623-29.
- Colle JG, Duguid JP, Fraser AG, Marimon BP, Simons A. Laboratory strategy in the diagnosis of infective syndromes. Mackie and McCartney Practical Medical Microbiology 14 (1996): 53-94.
- Toledo-Arana A, Valle J, Solano C, Arrizubieta MJ, Cucarella C, Lamata M et al. The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Environ. Microbiol 67 (2001): 4538-45.
- Stepanovic S, Vukovic D, Hola V, BONAVENTURA GD, Djukic S, Cirkovic I et al. Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by Staphylococci. Apmis 115 (2007): 891-99.
- Jia W, Li G, Wang W. Prevalence and antimicrobial resistance of Enterococcus species: a hospital-based study in China. J Environ Res Public Health 11 (2014): 3424-42.
- Vankerckhoven V, Van Autgaerden T, Vael C, Lammens C, Chapelle S, Rossi R et al. Development of a multiplex PCR for the detection of asa1, gelE, cylA, esp, and hyl genes in Enterococci and survey for virulence determinants among European hospital isolates of Enterococcus faecium.J. Clin. Microbiol 42 (2004): 4473-79.
- Moniri R, Ghasemi A, Moosavi SG, Dastehgoli K, Rezaei M. Virulence gene’s relationship with biofilm formation and detection of aac (6’)/aph (2”) in Enterococcus faecalis isolated from patients with urinary tract infection. Jundishapur J. Microbiol 6 (2013): e94137.
- Ch’ng JH, Chong KK, Lam LN, Wong JJ, Kline KA. Biofilm-associated infection by enterococci. Nature Reviews Microbiology 17 (2019): 82-94.
- Komiyama EY, Lepesqueur LS, Yassuda CG, Samaranayake LP, Parahitiyawa NB, Balducci I, Koga-Ito CY. Enterococcus species in the oral cavity: prevalence, virulence factors and antimicrobial susceptibility. PloS one 11 (2016): e0163001.
- Mohanty S, Behera B. Antibiogram Pattern and Virulence Trait Characterization of Enterococcus Species Clinical Isolates in Eastern India: A Recent Analysis. J Lab Physicians 14 (2022): 237-46.
- Shridhar S and Dhanashree B. Antibiotic Susceptibility Pattern and Biofilm Formation in Clinical Isolates of Enterococcus spp. Interdisciplinary perspectives on infectious diseases (2019).
- Zheng JX, Wu Y, Lin ZW, Pu ZY, Yao WM, Chen Z, et al. Characteristics of and virulence factors associated with biofilm formation in clinical Enterococcus faecalis isolates in China. Microbiol 8 (2017): 2338.
- Palmer KL, Kos VN, Gilmore MS. Horizontal gene transfer and the genomics of enterococcal antibiotic resistance. Curr. Opin. Microbiol 13 (2010): 632-9.
- Kashef M, Alvandi A, Hasanvand B, Azizi M and Abiri R. Virulence factor and biofilm formation in clinical Enterococcal isolates of the west of Iran. Jundishapur J Microbiol 10 (2017): e14379.
- Hashem YA, Yassin AS and Amin MA. Molecular characterization of Enterococcus spp. clinical isolates from Cairo, Egypt. Indian J Med Microbiol 33 (2015): 80-6.
- Chajecka-Wierzchowska W, Zadernowska A, Laniewska-Trokenheim L. Virulence factors of Enterococcus spp. presented in food. LWT-Food Sci Technol 75 (2017): 670-76.
- SIKDAR S, SADHUKHAN S, MAJUMDAR AK, BHUNIA S, SARKAR S, BHATTACHARJEE SG. Phenotypic Characterisation, Virulence Determination and Antimicrobial Resistance Pattern of Enterococcus Species Isolated from Clinical Specimen in a Tertiary Care Hospital in Kolkata. J. Clin. Diagnostic Res 15 (2021).
- Abdel-Gawad AR, Rezk GA, Mahmoud EA. Characterization of Enterococci isolated from intensive care unit (ICU); Distribution of virulence markers, virulence genes and antibiotic resistance pattern. Microbes and Infectious Diseases 2 (2021): 725-35
- Dworniczek E, Wojciech L, Sobieszczanska B & Seniuk A. Virulence of Enterococcus isolates collected in Lower Silesia (Poland). Scand J Infect Dis 37 (2005): 630–36.
- Banerjee T and Anupurba S. Prevalence of virulence factors and drug resistance in clinical isolates of Enterococci: A study from North India. J. pathog (2015).
- Di Rosa R, Creti R, Venditti M, D’Amelio R, Arciola CR, Montanaro L. Relationship between biofilm formation, the enterococcal surface protein (Esp) and gelatinase in clinical isolates of Enterococcus faecalis and Enterococcus faecium. FEMS Microbiol Lett 256 (2006): 145–50.
- Saffari F, Dalfardi MS, Mansouri S and Ahmadrajabi R. Survey for correlation between biofilm formation and virulence determinants in a collection of pathogenic and fecal Enterococcus faecalis Infect chemother 49 (2017): 176-83.
- Lee Y. Biofilm formation and antimicrobial resistance in Enterococcus. Infect chemother 49 (2017): 236-7.
- Jett BD, Huycke MM, Gilmore MS. Virulence of Enterococci. Microbiol Rev 7 (1994): 462-78.
- DeVuyst L, Moreno MF and Revets H. Screening for enterocins and detection of hemolysin and vancomycin resistance in Enterococci of different origins. J. Food Microbiol 84 (2003): 299-318.
- Han D, Unno T, Jang J, Lim K, Lee SN, Ko G, Sadowsky MJ, Hur HG. The occurrence of virulence traits among high-level aminoglycosides resistant Enterococcus isolates obtained from feces of humans, animals, and birds in South Korea. J Food microbiol 144 (2011): 387-92.
- Sienko A, Wieczorek P, Majewski P, Ojdana D, Wieczorek A, Olszanska D et al. Comparison of antibiotic resistance and virulence between biofilm-producing and non-producing clinical isolates of Enterococcus faecium. Acta Biochim Pol 62 (2015): 859-66
- Jankoska G, Trajkovska-Dokic E, Panovski N, Popovska-Jovanovska K and Petrovska M. Virulence factors and antibiotic resistance in Enterococcus faecalis isolated from urine samples. Prilozi 29 (2008): 57-66.


 Impact Factor: * 3.5
Impact Factor: * 3.5 Acceptance Rate: 71.36%
Acceptance Rate: 71.36%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 
