Transcriptome and SARS-CoV-2 Biological Network Directed Analysis for Better Therapeutic Development
Om Prakash Sharma1*, Ruchika Sharma2 and Vinay Chandil2
1Innoplexus AG, Frankfurter Str. 27, Eschborn, 65760 Germany
2Innoplexus Consulting Pvt. Ltd, 7th Floor, Midas Tower, Next to STPI Building, Phase 1, Hinjewadi Rajiv Gandhi, Infotech Park, Hinjewadi, Pune, Maharashtra 411057
*Corresponding author: Om Prakash Sharma, Innoplexus AG, Frankfurter Str. 27, Eschborn, 65760 Germany.
Received: 11 October 2023; Accepted: 20 October 2023; Published: 30 October 2023
Article Information
Citation: Om Prakash Sharma, Ruchika Sharma and Vinay Chandil. Transcriptome and SARS-CoV-2 Biological Network Directed Analysis for Better Therapeutic Development. Fortune Journal of Health Sciences. 6 (2023): 403-421.
View / Download Pdf Share at FacebookAbstract
Background: It has been almost 3.5 years since the first SARS-CoV-2 virus was first reported in the city of Wuhan. While the FDA has approved a number of drugs for Covid-19, the presence of the disease and its symptoms underscores the continued demand for an improved treatment option to effectively address the existing challenges. In this study, our goal is to identify pivotal protein targets, strongly correlated across lung, blood, and peripheral blood mononuclear cell (PBMC) transcriptomics datasets, to suggest promising targets for comprehensive therapeutic development across multiple tissues.
Methods: Transcriptomics datasets were retrieved from Geo Omnibus (GEO). We use relevant datasets to identify the most significant and differentially expressed genes and integrated them into a Research graph called CovInt (a network of Covid-19) that includes all biological molecules associated in the network with their directionalities collected from publicly available and patient-derived multi-omics datasets from millions of unstructured and structured datasets such as publications, patents, grants, preclinical and clinical reports. CovInt utilizes powerful traversal, clustering and centrality algorithms to identify key connections in the pathophysiology of the disease and its treatments.
Results: Leveraging 3M+ connections, important interactions among key 42 drugs, 962 biological processes and molecular functions, 926 pathways, 897 phenotypes, 7103 proteins, 61 tissues were identified. This narrowed interactome was explored further using PageRank, lovain detection & strongly connected components (SSC) algorithms. In our analysis, 63 strongly connected communities were identified which gives us an understanding of hidden underlying mechanisms. We further explored this network to identify and triangulate the key proteins, metabolic pathways and associated risk factors that can regulate moderate to severe Covid-19 infections.
Conclusions: Our study suggests that CREB3L1, SOX2, UBR4, FLNC, ITPA, DLG3, ING4, TECR, NADH, SMAD, HUWE1, DDX17, CREBBP, RELA, HPSE, TRIM33, TNFSF13B are the key regulator proteins in PBMC, Blood and Lungs in Covid19 patients. These proteins are involved in ER-stress, cytokine signaling, T-Cell activation, Activation of NLRP3 Inflammation by SARS-CoV-2, JAK-STAT, IL-4, IL-13 pathways, MAPK signaling pathways, Activation of NMDA receptor & postsynaptic events and TGF-β signaling pathways. This set of proteins needs to be further investigated in experimental studies for better therapeutic design of Covid-19.
Keywords
<p>Covid-19; transcriptomic analysis; SARS-CoV-2 targets; differentially expressed genes; network analysis; target engagement; perturbed pathway</p>
Article Details
1. Introduction
The ongoing COVID-19 infection is still a major concern for most of the country and has made a serious impact on worldwide public health. The world has seen a number of mutational variants and as a consequence huge mortality and cases of hospitalization. The virus mostly affects the human breathing and immune system which ultimately leads to respiratory distress syndrome (ARDS), cardiac issues [1], multi-organ failure and eventually death [2]. The SARS-CoV-2 belongs to the Betacoronavirus group and in the past few years, we have seen how frequently this virus is constantly changing through multiple mutations and emerging as a new therapeutic challenge to the world by showing resistance to the available therapeutics and prevention vaccines [3 – 5]. We believe there are high chances that in future as well these viruses will get mutated and may cause serious health causes. Since December 2019, the virus has mutated significantly and transmitted rapidly all over the world. A number of mutations have been reported to date for SARS-CoV-2 which is classified by WHO and CDC in four different types (1) Variant Being Monitored (VBM) (2) Variant of Interest (VOI) (3) Variant of Concern (VOC) and (4) Variant of High Consequence (VOHC). As per European Centre for Disease Prevention and Control (ECDC), as of 17th March’2022, 1,145,785, alpha and 40,534 Beta, 4,226,252 Delta and 2,277,587 Omicron variants of concerned genomes have been isolated and processed [6]. The Variant of Concern is a class of variants that are spreading fast and making a severe impact on public health. Currently, the Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2) and Omicron (B.1.1.529)7 variants are the only variants that fall in this group.
To date, a total of 5,401 interventional clinical studies are reported on clinicaltrial.gov and out of which 364 studies from Phase3 and 100 clinical studies from Phase-4 are completed. However, only limited therapeutics are approved so far to treat mild to moderate SARS-CoV-2 infection with huge unmet needs [7, 8] and treatment benefits with the frequent mutation are still a matter of concern. Due to the heterogeneity among various populations and diversification of SARS-CoV-2 mutations, it is an alarming situation to understand the crucial target engagement of the disease and uncover various underlying disease pathophysiology based on disease severity, stage, tissues, ages and patient populations. Several researchers have reported the importance of transcriptomic signature in response to infection [9]. Therefore, the transcriptome signature of SAR-CoV-2 infection could be one of the important parameters to identify the most crucial panel of targets engaged in the pathophysiology and severity of the disease. In this current research work, 15 high throughput transcriptomics datasets were selected from the Gene Expression Omnibus (GEO) database. These datasets were further analyzed to predict the most significant DEGs that can differentiate between healthy and disease patients based on Blood, PBMC and Lung tissues datasets. Later, these genes were further evaluated through the in-house built Covid-19 interacting network (CovInt) to investigate their importance in SARS-CoV-2 pathophysiology. In order to identify these crucial regulatory genes, we leveraged various network scoring such as page rank algorithms and the popularity of molecules within the network. The present study aimed to identify the most significant panel of gene sets which can be used for better therapeutic development against Covid-19 infections.
2. Materials and Methods
2.1 Transcriptomics data collection and validation
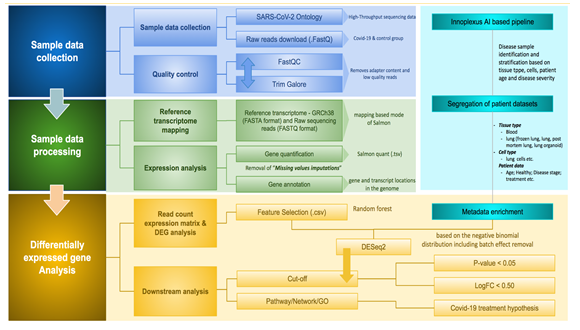
Gene Expression analysis was carried out starting from the raw FastQ sequencing data downloaded from Gene Expression Omnibus (GEO) database [10]. We searched using “Severe acute respiratory syndrome coronavirus 2 [Organism] OR Covid-19 [All Fields]) OR Severe acute respiratory syndrome coronavirus 2 [Organism] OR covid19 [All Fields] OR Severe acute respiratory syndrome coronavirus 2 [Organism] OR SARS-CoV-2 [All Fields]) AND "Homo sapiens"[porgn] AND "gse"[Filter]” on 3rd Jan 2022 in the GEO database for SARS-CoV-2 associated expression datasets which results in “311” hits. Later, we applied filters on study types using Expression profiling by array and Expression profiling by high throughput sequencing (HTS) which results in 300 studies. These series were downloaded in .txt format from GEO omnibus data portal. The obtained file was converted into .csv format for further validation and classification in the Linux terminal using the below-mentioned syntax
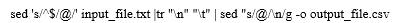
Later, we used our in-house developed AI-based tool to stratify and label samples based on their tissue types, study types and sub-indications. This in-house developed model also removes studies with less than 3 samples, samples with nonspecific disease names, studies with only healthy data, superseries and other studies such as snRNA-seq, single-cell, and missing SRA id datasets and classified as irrelevant/insignificant studies. The remaining relevant studies were further analyzed for differentially expressed genes. All relevant studies were carried forward for further analysis.
2.2 Transcriptomics data analysis
Relevant expression datasets were further processed for further quality control using Trim-galore [11] which trims off all low-quality bases from the 3’ end of the reads before adapter removal. In the next step, all adapters and short sequences (20 bp) from the 3’ end of reads were filtered. Estimating transcript abundance from RNA-seq reads is a fundamental and crucial step in transcriptomic analysis. We used Salmon [12] as a tool to quantify transcripts based on the reference transcriptome build GRCh38 downloaded from NCBI and indexed using Salmon. The number of reads per transcript obtained for each sample from previous steps was directly used as an input to calculate Differentially Expressed Genes (DEGs). DEG analysis was carried out by using the DESeq2 [13] Bioconductor package v1.34.0, which calculated Log2FC (log 2-fold change) values per gene including p-values, adjusted p-value and base mean values. We applied a cut-off p-value for < 0.005 and Log2FC value > 1.5 for further analysis.
2.3 Enrichment of DEGs and development of CoVint network for molecular connections
Identified differentially expressed gene from the previous study was further used for the enrichment of associated pathways, molecular connections and disease-protein-associated relevant articles through Ontosight® Discover [14, 15] and Ontosight® Explore (https://www.innoplexus.com/blog/accelerate-your-research-and-discovery-in-life-sciences/). The overall transcriptomics analysis process is shown in Figure 1.
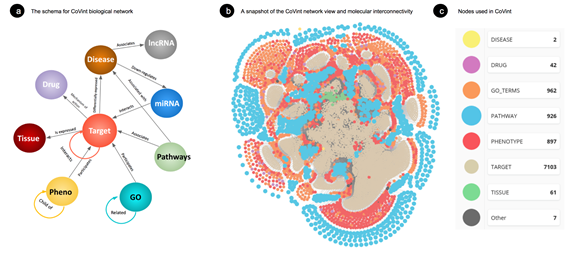
Ontosight Explore® [16, 17] consist of 4.2 M+ chemicals and drug, 250 K+ disease, 560 K+ proteins, 6 K+ pathways and 39 K+ number of molecular connections extracted from various literature and publicly available curated database including Covid-19 pathways from WikiPathways [18] and build a comprehensive SARS-CoV-2 protein-protein interaction network called CoVint (Covid Interactome) by maintaining their types of relationships and directionality of reactions in a neo4j v4.3.3 graph database. The overall schema of CovInt has been shown in Figure 2a. In this current study, Ontosight Discover® and Explore® were used to identify the molecular connections of prioritized DEGs and their significance in Covid-19 pathophysiology.
Here, CoVint consists of 39 K+ molecular connections associated with SARS-CoV-2 (Figure 2b). These connections are then further traversed using the neo4j browser and visualized using neo4j’s bloom application. We superimposed expression values of each DEG in the CoVint biological network to determine the flow of expression and identify important perturbations in the network. This helped us in taking into account the effect of expression change from healthy to disease on other molecular pathways and entities.
(b) A SARS-CoV-2 differentially expressed gene and its biological connection. Here, the yellow color demonstrates covid19 disease, while related biological pathways are highlighted in blue color, clinical and biological function is shown in red colors, interacting partner proteins are shown in light brown color, tissues are in green and other entities are shown in grey color.
To prioritize DEGs We used a Hyperlink-Induced Topic Search (HITS) algorithm to check the flow of expression from a DEG in the network which helped in identifying the authority DEGs that have a greater influence. The developed network is presented in Figure 2b.
2.4 Single pathway network (SPN) analysis and GO enrichment
We analyzed each covid19 pathway (P) by superimposing its protein associations (g1, g2...gn) with their interactions such that
PI(g(k)) = [i(k)(1), i(k)(2)...i(k)(m)] where k<=n
Thus the imposed pathway network consisted of 1 pathway node (P), L = union of (g, PI(g)) protein nodes and (P)-[:Associated with]->(L), (g)-[:Interacts]-(PI(g)) relations. This concluded the generation of our pathway network. Then we mapped the differentially expressed gene on this network to find the importance and influence of this pathway (W(p)) using a greedy influence maximization approach where a group of proteins that when expressed may induce an effect of the pathway.
In order to understand the biological functions of predicted DEGs, we performed Gene Ontology (GO) [19]. We first downloaded all GO datasets (http://geneontology.org/docs/downloads/) with their associated genes, later we performed statistical analysis to enrich Gene ontology for the identified DEGs. Identified genes were classified into GO categories as a biological process (BP), molecular function (MF) and cellular component (CC). Top enriched GO was identified based on the p < 0.05 as the cut-off criterion and various graphs were generated to understand the overlapped genes and Gene ontologies involved in disease pathophysiology. We further used this information to triangulate this information with the CoVint network and evaluated the top marker using published scientific reports.
3. Results
3.1 Transcriptomics data collection and analysis
In order to perform transcriptomics analysis, we first queried the GEO Omnibus database to identify the most relevant datasets for our study. It results in 311 hits for our given query against COVID-19. The details of various types of datasets have been shown in Table 1.
Table 1: SARS-CoV-2 associated different datasets
|
# |
Study type |
Studies |
|
1 |
Expression profiling by array |
12 |
|
2 |
Expression profiling by high throughput sequencing |
300 |
|
3 |
Genome binding/occupancy profiling by array |
0 |
|
4 |
Genome binding/occupancy profiling by high throughput sequencing |
7 |
|
5 |
Methylation profiling by array |
1 |
|
6 |
Methylation profiling by high throughput sequencing |
1 |
|
7 |
Non-coding RNA profiling by array |
0 |
|
8 |
Non-coding RNA profiling by high throughput sequencing |
4 |
However, upon validation and stratification of relevant samples, we remained with 16 studies which we were able to include in our further study. The remaining 295 studies were excluded from our further analysis (Supplementary Table 1). In total, we identified 12 high throughput sequencing studies and 4 Microarray studies. 2 out of the 4 Microarray studies were done on immune-specific probes, therefore due to missing data we discarded those studies (Table 1). Among the 12 HTS studies, 5 studies were from Blood samples, 2 studies were from Peripheral Blood Mononuclear Cells (PBMCs), 2 studies were from Lung Tissue with a significant number of studies which we carried forward for further analysis. In total, we identified 201 healthy volunteers and 575 disease samples where we had a significant amount of data to generate confident results. Single studies from other sample sources such as Nasal Swabs, Brain, and Platelets were discarded due to a limited number of experiments (Table 2).
Table 2: Characteristics of High-throughput studies datasets considered in this study
|
# |
Study type |
Studies |
GSE series |
Healthy |
Disease |
Total Sample |
Considered for study? |
|
1 |
PBMC |
2 |
GSE166253 |
16 |
10 |
94 |
Yes |
|
GSE152418 |
34 |
34 |
Yes |
||||
|
2 |
Blood |
5 |
GSE185557 |
18 |
21 |
39 |
Yes |
|
GSE166190 |
16 |
82 |
98 |
Yes |
|||
|
GSE169687 |
14 |
138 |
152 |
Yes |
|||
|
GSE161731 |
19 |
58 |
77 |
Yes |
|||
|
GSE161777 |
13 |
14 |
27 |
Yes |
|||
|
3 |
Lung |
2 |
GSE151764 |
16 |
34 |
50 |
Yes |
|
GSE150316 |
15 |
83 |
98 |
Yes |
|||
|
4 |
Nasal Swabs |
1 |
GSE166530 |
5 |
36 |
41 |
No |
|
5 |
Brain |
1 |
GSE179923 |
6 |
1 |
7 |
No |
|
6 |
Platelets |
1 |
GSE176480 |
10 |
8 |
18 |
No |
Identified samples were further normalized using Limma-remove batch effect package [20] and visualization was done using PCA plot. Since we used DESeq2 for getting DEGs, where expression values are normalized using the median-ratios method, we used the Limma package to analyze the data batches in detail and how the data looks after removing the batch effect.
3.2 Identification of DEGs
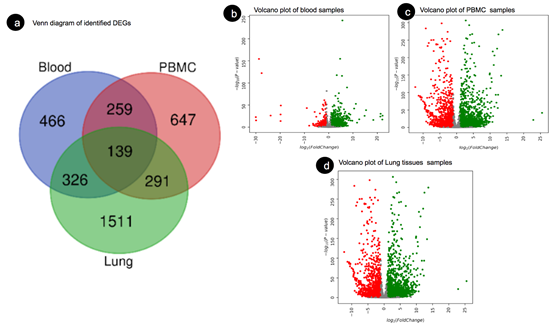
Based on the cut-off criteria of log2 FC > 1.5 for upregulated and < -1.5 for downregulated and adj P-value < 0.05. We identified 925, 1336 and 50 upregulated genes and 265, 938 and 2217 downregulated genes in Blood, PBMC and Lung respectively and a total of 139 overlapping DEGs were identified as shown in Figure 3a obtained from the Lung, Blood and PBMC samples. The DEGs identified for each of the tissues separately, are represented through volcano plots as shown in Figures 3b, 3c and 3d respectively. The list of identified DEGs can be found here as Supplementary File 2.
Figure 3: (a) Venn diagram of identified DEGs, 139 overlapped DEGs from Blood, PBMC and Lung samples. (b) Volcano plot of identified DEGs from blood tissue samples (C) Volcano plot of PBMC samples (d) Volcano plot of Lung tissue samples. Here, the red color demonstrates downregulated genes, and the green color demonstrates upregulation of genes.
The expression level of each sample is shown in the Volcano plot in Figure-3b, 5c and 5d. Moreover, the heatmap of DEGs demonstrates that these DEGs could distinguish the control and other samples.
3.3 Gene Functional Enrichment from Top Degs of Blood, PBMC and Lung Tissues
3.3.1 Blood tissues
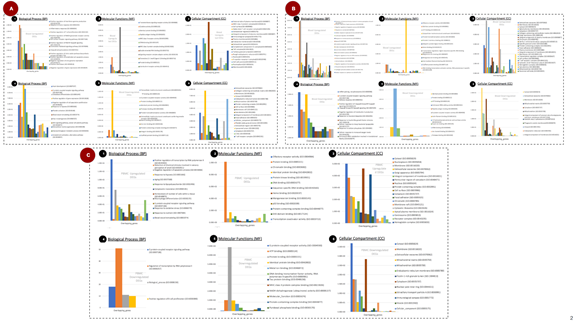
GO enrichment of top upregulated gene suggests that Detection of chemical stimulus involved in sensory perception of smell (GO:0050911), Positive regulation of gene expression (GO:0010628), Negative regulation of apoptotic process (GO:0043066), Response to nutrient (GO:0007584), Cellular response to glucose stimulus (GO:0071333) (Figure 4A.1), whereas, GO enrichment of top downregulation gene suggests that mRNA splicing, via spliceosome (GO:0000398), G protein-coupled receptor signaling pathway (GO:0007186), Defense response (GO:0006952), Positive regulation of I-kappaB kinase/NF-kappaB signaling (GO:0043123), I-kappaB kinase/NF-kappaB signaling (GO:0007249) are the major biological processes (Figure 4A.4).
Olfactory receptor activity (GO:0004984), Identical protein binding (GO:0042802), ATP binding (GO:0005524), Extracellular matrix structural constituent (GO:0005201), Protein homodimerization activity (GO:0042803) are the top five molecular functions of identified upregulated DEGs also demonstrated in Figure 4A.2. and Identical protein binding (GO:0042802), Protein binding (GO:0005515), ATP binding (GO:0005524), Aminoacyl-tRNA editing activity (GO:0002161), Peptide antigen binding (GO:0042605) are the top molecular functions which influenced due to downregulation of identified DEGs (Figure 4A.5).
Similarly, Extracellular exosome (GO:0070062), Membrane (GO:0016020), Cytosol (GO:0005829), Apical plasma membrane (GO:0016324), Cytoplasm (GO:0005737) are the top cellular components in blood tissue from upregulated DEGs (Figure 4A.3) and Cytosol (GO:0005829), Extracellular exosome (GO:0070062), Membrane (GO:0016020), Mitochondrial matrix (GO:0005759), Cytoplasm (GO:0005737) are the top cellular components influenced by downregulated DEGs (Figure 4A.6).
3.4 Peripheral blood mononuclear cell (PBMC)
Top biological processes enriched from upregulated DEGs are PBMC are Positive regulation of transcription by RNA polymerase II (GO:0045944), Detection of chemical stimulus involved in sensory perception of smell (GO:0050911), Negative regulation of apoptotic process (GO:0043066), Response to hypoxia (GO:0001666), Aging (GO:0007568) have been also shown in Figure 4B.1 and downregulation of DEGs G-protein-coupled receptor signaling pathway (GO:0007186), Regulation of transcription by RNA polymerase II (GO:0006357), Biological process (GO:0008150), Positive regulation of B cell proliferation (GO:0030890) have been shown in Figure 4B.4. Olfactory receptor activity (GO:0004984), Protein binding (GO:0005515), Chromatin binding (GO:0003682), Identical protein binding (GO:0042802), Protein kinase binding (GO:0019901) were top molecular functions from upregulated DEGs (Figure 4B.2) whereas G-protein-coupled receptor activity (GO:0004930), Protein binding (GO:0005515), ATP binding (GO:0005524), Identical protein binding (GO:0042802), Metal ion binding (GO:0046872) are from Downregulated DEGs (Figure 4B.5)
Similarly, Cytosol (GO:0005829), Nucleoplasm (GO:0005654), Membrane (GO:0016020), Extracellular exosome (GO:0070062), Golgi apparatus (GO:0005794) are the top cellular component enriched from the upregulated PBMC genes (Figure 4B.3). Whereas Cytosol (GO:0005829), Membrane (GO:0016020), Extracellular exosome (GO:0070062), Mitochondrial matrix (GO:0005759), Mitochondrion (GO:0005739) are the top enriched CC (Figure 4B.6) from top downregulated DEGs. We observed in both types of DEGs cytosol, the Extracellular exosome and Membrane are the most important CC that was influenced (Figure 4B).
3.5 Lung tissues
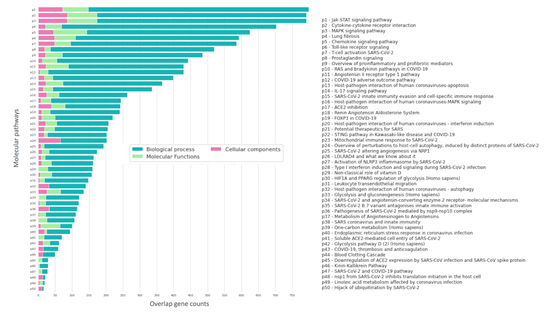
GO enrichment from top downregulated DEGs from lung tissues revealed Positive regulation of interferon-gamma production (GO:0032729), Adaptive immune response (GO:0002250), Inflammatory response (GO:0006954), Positive regulation of T cell proliferation (GO:0042102), Positive regulation of NMDA glutamate receptor activity (GO:1904783) are the major biological processes (Figure 4C.1) and Heart development (GO:0007507), G protein-coupled receptor signaling pathway (GO:0007186), Osteoblast differentiation (GO:0001649), Positive regulation of gene expression (GO:0010628), Negative regulation of cell population proliferation (GO:0008285) are the top BP from downregulated DEGs (Figure 4C.4). Similarly, the top cellular components (CC) from the upregulated genes are the External side of the plasma membrane (GO:0009897), MHC class II protein complex (GO:0042613), Cell surface (GO:0009986), Alpha-beta T cell receptor complex (GO:0042105), Extracellular region(GO:0005576) (Figure 4C.2) and Extracellular matrix structural constituent (GO:0005201), ATP binding (GO:0005524), G protein-coupled receptor activity (GO:0004930), Protein binding (GO:0005515), Identical protein binding (GO:0042802) for downregulated DEGs (Figure 4C.6). External side of the plasma membrane (GO:0009897), MHC class II protein complex (GO:0042613), Cell surface (GO:0009986), Alpha-beta T cell receptor complex (GO:0042105), Extracellular region (GO:0005576) (Figure 4C.3) are top CC from upregulated Genes and Extracellular exosome (GO:0070062), Collagen-containing extracellular matrix (GO:0062023), Membrane (GO:0016020), Cytosol (GO:0005829), Endoplasmic reticulum lumen (GO:0005788) are top CC from downregulated DEGs (Figure 4C.6). In order to understand the role and involvement of identified genes and enriched BP, MF and CC, we performed a pathway over-representation study (Figure 9). Our study revealed that the Jak-STAT signaling pathway [21], Cytokine-Cytokine receptor interactions [22], MAPK signaling pathway [23], Lung Fibrosis [24–26], Chemokine signaling pathway [24], Toll-like receptor signaling are the major pathways that were represented by identified DEGs and GO analysis. These pathways are well known and reported previously to play a crucial role in SARS-CoV-2-associated pathophysiology (Figure 5).
Figure 5: Enriched molecular pathways from the top 15 GO of BP, MF and CC. Here, XX color represents Biological processes, YY represents Cellular components and ZZ color represents Molecular functions. The bar of these pathways was represented through the overlap of identified GO genes with molecular pathways.
Network analysis
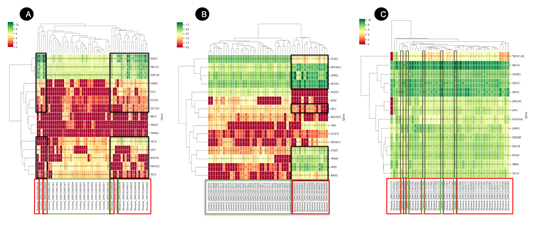
The identified DEGs were further explored in the CovInt network to understand their biological connections and their importance in the sub-network. Functional and CoVint network analysis revealed MAGED1, FBXO7, ATAD3A, TECR, CCDC8, CDC14A, TERF2IP, FBXW7, RNF123, CAND1, USP7, SMC4, HIF1A, TRIM63, CUL1 as the top 15 hub genes from blood tissues (Figure 6A) and IKBKG, CTDP1, TNIK, BTRC, FBXW11, STUB1, NCOR2, LRRK2, HPSE, FHOD1, CCDC8, ARFGEF2, TRRAP, NDUFAF1, CBY1 (Figure 6B) from PBMC.
Figure 6: Heatmap of DEGs in (A) Blood (B) PBMC and (C) Lung sample. Each row represents a single gene, and each column represents a sample. Disease-associated samples are highlighted in red boxes whereas healthy samples are demonstrated in green boxes. The color scale shows the relative gene expression in certain slides. Green represents high relative expression levels and red indicates low relative expression levels.
Whereas, PLEKHA4, CHCHD2, SUPT16H, LRRK2, PTRH2, STUB1, CCDC8, UBQLN1, SMC3, UBQLN2, MAP1LC3B, DOLK, AFG3L2, RBM8A, TBK1 are the top 15 hub genes from lung tissues (Figure 6C). We identified multiple clusters and sub-networks of various SARS-CoV-2-associated genes which were further prioritized based on the LogFc value, Network Score, target overlap count with covid-19 pathways and p-value score parameters. We estimated the relative rank for each parameter using Microsoft excel as shown below -
Relative ranking = RANK($A2,$A$2:$A$N)
Here, A represents the name of the column and “N” represents the number of rows
And named as “Relative gene expression ranking” for LogFC, “Covid-19-associated pathway” for covid-19 pathways, “Relative network score” for Network Score and estimated a consolidated score using the below-mentioned formula where p-value was used as a qualifying parameter.
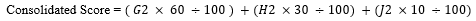
Here, G2 stands for CovInt network Rank; H2 stands for relative Expression Ranking and J2 stands for relative covid-19 associated Pathway ranking. Based on relative ranking the top 50 relevant DEGs from Blood have been shown in Table 3, PBMC in Table 4 and Lung tissues in Table 5.
Table 3: The DEGs of merged datasets from the blood sample with the applied criteria of p-value <0.05 and |log2FC| > 1.5, network score and directionality of regulations. The table is sorted based on the consolidated ranking score.
|
Blood tissues |
|||||||
|
# |
Gene Symbol |
Gene Name |
Network Score |
Gene Expression |
Direction |
p-value |
Relative ranking |
|
1 |
SLC25A13 |
Solute carrier family 25 member 13 |
1.254 |
-4.706 |
down |
2.60E-02 |
1 |
|
2 |
FBXW7 |
F-box and WD repeat domain containing 7 |
1.866 |
-3.325 |
down |
4.30E-06 |
2 |
|
3 |
NOD2 |
Nucleotide binding oligomerization domain containing 2 |
0.969 |
-3.862 |
down |
2.85E-08 |
3 |
|
4 |
CREB3L1 |
cAMP responsive element binding protein 3 like 1 |
1.091 |
6.338 |
up |
3.93E-10 |
4 |
|
5 |
CDC14A |
cell division cycle 14A |
2.063 |
5.07 |
up |
9.42E-16 |
5 |
|
6 |
SMC4 |
Structural maintenance of chromosomes 4 |
1.487 |
5.147 |
up |
5.35E-05 |
6 |
|
7 |
CFL2 |
Cofilin 2 |
1.276 |
-2.58 |
down |
4.21E-04 |
7 |
|
8 |
MAGED1 |
MAGE family member D1 |
8.364 |
-2.206 |
down |
5.36E-04 |
8 |
|
9 |
ITPA |
Inosine triphosphatase |
0.758 |
-3.788 |
down |
3.05E-08 |
9 |
|
10 |
DLG3 |
Discs large MAGUK scaffold protein 3 |
0.678 |
-4.79 |
down |
7.53E-04 |
10 |
|
11 |
ING4 |
Inhibitor of growth family member 4 |
0.873 |
-2.853 |
down |
1.10E-02 |
11 |
|
12 |
TECR |
Trans-2,3-enoyl-CoA reductase |
2.586 |
-2.093 |
down |
6.00E-07 |
12 |
|
13 |
RELA |
RELA proto-oncogene, NF-kB subunit |
1.168 |
4.78 |
up |
2.82E-06 |
13 |
|
14 |
CDH1 |
Cadherin 1 |
0.829 |
5.59 |
up |
3.78E-06 |
14 |
|
15 |
ATAD3A |
ATPase family AAA domain containing 3A |
2.651 |
-1.959 |
down |
4.50E-03 |
15 |
|
16 |
FBXO7 |
F-box protein 7 |
8.101 |
4.215 |
up |
2.20E-30 |
16 |
|
17 |
ACAD8 |
Acyl-CoA dehydrogenase family member 8 |
0.807 |
-2.499 |
down |
5.51E-03 |
17 |
|
18 |
RASSF1 |
Ras association domain family member 1 |
0.605 |
-3.601 |
down |
1.69E-31 |
18 |
|
19 |
SOX2 |
SRY-box transcription factor 2 |
0.783 |
5.018 |
up |
9.94E-10 |
19 |
|
20 |
IGSF8 |
Immunoglobulin superfamily member 8 |
0.561 |
-4.588 |
down |
3.06E-09 |
20 |
|
21 |
MID2 |
Midline 2 |
0.911 |
-2.177 |
down |
1.53E-02 |
21 |
|
22 |
CUL1 |
Cullin 1 |
1.39 |
4.193 |
up |
1.26E-06 |
22 |
|
23 |
SP110 |
SP110 nuclear body protein |
0.493 |
-19.754 |
down |
3.36E-29 |
23 |
|
24 |
EHMT1 |
Euchromatic histone lysine methyltransferase 1 |
0.66 |
-2.737 |
down |
4.53E-07 |
24 |
|
25 |
NODAL |
Nodal growth differentiation factor |
0.8 |
4.751 |
up |
1.38E-07 |
25 |
|
26 |
PPAN |
Peter pan homolog |
0.899 |
-2.07 |
down |
2.08E-04 |
26 |
|
27 |
PRDM16 |
PR/SET domain 16 |
1.155 |
4.182 |
up |
1.86E-09 |
27 |
|
28 |
FAM118B |
Family with sequence similarity 118 member B |
0.466 |
-5.761 |
down |
6.92E-35 |
28 |
|
29 |
IFT172 |
Intraflagellar transport 172 |
0.491 |
-3.891 |
down |
4.08E-02 |
29 |
|
30 |
SYNC |
Syncoilin, intermediate filament protein |
0.475 |
7.565 |
up |
2.75E-90 |
30 |
|
31 |
UHRF1 |
Ubiquitin like with PHD and ring finger domains 1 |
0.614 |
5.168 |
up |
2.17E-08 |
31 |
|
32 |
SH3GLB1 |
SH3 domain containing GRB2 like, endophilin B1 |
1.361 |
3.857 |
up |
3.10E-16 |
32 |
|
33 |
GANAB |
Glucosidase II alpha subunit |
0.465 |
-4.146 |
down |
8.93E-09 |
33 |
|
34 |
TAF15 |
TATA-box binding protein associated factor 15 |
1.058 |
-1.779 |
down |
1.74E-61 |
34 |
|
35 |
PTK2B |
Protein tyrosine kinase 2 beta |
0.576 |
5.093 |
up |
2.11E-32 |
35 |
|
36 |
HSPB8 |
Heat shock protein family B (small) member 8 |
1.148 |
3.86 |
up |
8.53E-06 |
36 |
|
37 |
DIABLO |
Diablo IAP-binding mitochondrial protein |
1.15 |
-1.59 |
down |
7.20E-03 |
37 |
|
38 |
PARK7 |
Parkinsonism associated deglycase |
0.715 |
-1.97 |
down |
7.36E-23 |
38 |
|
39 |
BIRC7 |
Baculoviral IAP repeat containing 7 |
0.806 |
-1.829 |
down |
2.62E-02 |
39 |
|
40 |
RUFY1 |
RUN and FYVE domain containing 1 |
0.422 |
-6.191 |
down |
5.96E-06 |
40 |
|
41 |
CLUAP1 |
Clusterin associated protein 1 |
1.019 |
3.774 |
up |
7.16E-06 |
41 |
|
42 |
PPIE |
Peptidylprolyl isomerase E |
0.631 |
-2.022 |
down |
1.15E-04 |
42 |
|
43 |
KCTD3 |
Potassium channel tetramerization domain containing 3 |
1.34 |
3.435 |
up |
1.59E-02 |
43 |
|
44 |
PPT1 |
Palmitoyl-protein thioesterase 1 |
0.45 |
5.673 |
up |
7.61E-242 |
44 |
|
45 |
BRD7 |
Bromodomain containing 7 |
0.513 |
4.73 |
up |
3.79E-155 |
45 |
|
46 |
UBR4 |
Ubiquitin protein ligase E3 component n-recognin 4 |
0.443 |
-2.879 |
down |
8.39E-13 |
46 |
|
47 |
FLNC |
Filamin C |
0.605 |
4.328 |
up |
4.34E-07 |
47 |
|
48 |
AGAP3 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
0.419 |
-3.2 |
down |
3.07E-07 |
48 |
|
49 |
MYO5B |
Myosin VB |
0.802 |
3.727 |
up |
6.06E-13 |
49 |
|
50 |
EFEMP1 |
EGF containing fibulin extracellular matrix protein 1 |
0.395 |
7.234 |
up |
2.34E-03 |
50 |
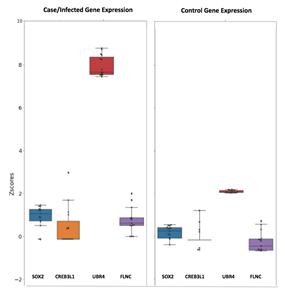
Transcriptomics profile analysis of Whole Blood, PBMC and lung suggested dysregulation of CREB3L1, SOX2, UBR4 and FLNC transcription factor (Figure 7). Several investigators has reported dysregulation of CREB3L1 in ER stress during the Covid-19 infections [27, 28] whereas SOX2 is known to be a bronchus progenitor marker gene [29–31] that is involved in cytokine-mediated signaling pathway [32] and play an important role in lung fibrosis [33, 34]. Another common transcription factor identified from this analysis is UBR4 which is an E3 ubiquitin ligase and was found to be involved in the membrane morphogenesis autophagy process during the cytokine storm [35]. In our study, we found UBR4 as significantly dysregulated in Covid-19 infections and can be used as a potential therapeutic target for Covid-19. UBR4 is previously reported as an important protein to be involved in the transportation of viral glycoproteins to the cell membrane and promotes replication of Influenza A virus [36]. Our study revealed a similar role of UBR4 in Covid-19 infections.
From blood tissue transcriptomics analysis, we identified Inosine Triphosphatase (ITPA) as one of the top transcription factors associated that are downregulated by -3.788 FoldChain as compared to a healthy patient. Multiple mutations are already reported in these genes such as rs1161447593, rs12980275 and rs8099917 which leads to a certain reduction in the ITPA expression associated with Hepatitis C virus response [37]. The role of ITPA can be further explored in SARS-CoV-2 infections. The second genetic marker is DLG3 which is found to be downregulated through our analysis with a Log2FC value of -4.790. DLG3 is recognized by SARS-CoV-2, E protein, however, its role still needs to be explored further [38]. Inhibitor of growth family member 4 (ING4) is another interesting transcription factor that is downregulated (-2.853) in the disease samples. It interacts with the CXCL8 gene and regulates cytokine during the cytokine storm inflammatory response.
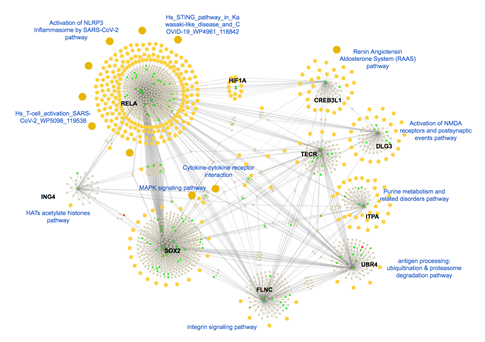
The expression of Trans-2,3-Enoyl-CoA Reductase (TECR) is downregulated (-2.093) in disease conditions. Activator of the TECR gene could be an interesting therapeutic target against Covid-19. RELA is another important key transcription factor that comes out from our analysis which is significantly upregulated (4.780) during the SARS-CoV-2 infection. RELA initiates TLR4 mediated NF-κB signaling pathway to activate IL-8 expression and thus regulates the IFN response [39]. Interestingly, upon investigation, we identified that these molecules are highly connected to each other and involved in various covid-19 infection-associated pathways such as Activation of NLRP3 inflammation, Renin-angiotensin Aldosterone system, Cytokine-cytokine receptor, MAPK signaling and Integrin signaling pathways (Figure 8).
Figure 8: Molecular connections of the top dysregulated gene from the Blood sample. Here, upregulated proteins are shown in the green bubbles and down-regulated proteins are shown in red color and neutral genes are demonstrated in gray color. All biological pathways are highlighted in yellow color. Covid-19 associated pathways are labeled using blue text to differentiate the sub-clustered and associated genes.
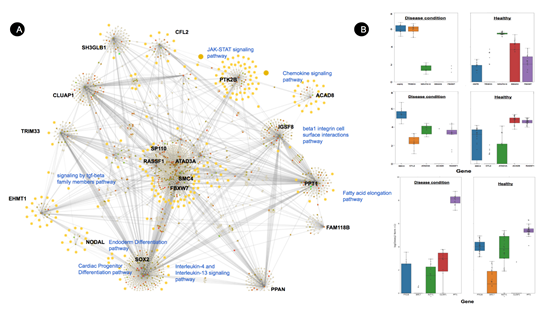
Transcriptomics analysis from PBMC samples revealed HPSE is generally upregulated during Covid-19 with Log2FC of 7.315, it is involved in pro-inflammatory glycocalyx generation that promotes chemokines, cytokines, and leukocytes binding to the endothelial cell surface. Inhibition of HPSE restricts HPSE activity and benefits Covid-19 patients [40] and other lung-associated diseases [41].
Tripartite Motif Containing 33 (TRIM33) is another interesting transcription factor that is found to be overexpressed (6.609) in our study. It regulates the proinflammatory function of Th17 cells [42]. Th17 cells are known to play an important role in COVID-19 pathogenesis by triggering cytokine cascade and inducing Th2 responses during the infection which leads to Treg cell suppression43,44.
Figure 9: (A) Molecular connections of the top dysregulated gene from PBMC sample. Here, upregulated proteins are shown in the green bubbles and down-regulated proteins are shown in red color and neutral genes are demonstrated in grey color. All biological pathways are highlighted in yellow color. Covid-19 associated pathways are labeled using blue text to differentiate the sub-clustered and associated genes. (B) Dysregulated genes from PBMC samples compared to healthy volunteers.
However, two important genes NADH and SMAD were identified as crucial hub genes from network analysis that can be activated for better therapeutic effects. Firstly, NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 12 (NDUFA12) which is an accessory subunit of the mitochondrial membrane respiratory chain NADH dehydrogenase (Complex I) is found to be downregulated with a -6.035 FoldChain during the SARS-COV-2 viral infection sample from our study. Secondly, SMAD proteins are down-regulated in our study -8.572, which is known to be involved in TGF-β signaling pathways and pulmonary fibrosis45.
Table 4: The DEGs of merged datasets from PBMC sample with the applied criteria of p-value <0.05 and |log2FC| > 1.5, network score and directionality of regulations
|
PBMC Tissues |
|||||||
|
# |
Gene Symbol |
Gene Name |
Network Score |
Gene Expression |
Direction |
p-value |
Relative ranking |
|
1 |
FHOD1 |
Formin homology 2 domain containing 1 |
4.261 |
-10.042 |
down |
1.58E-75 |
1 |
|
2 |
HPSE |
Heparanase |
4.538 |
7.315 |
up |
3.11E-42 |
2 |
|
3 |
MAD2L2 |
Mitotic arrest deficient 2 like 2 |
3.446 |
-6.16 |
down |
5.81E-184 |
3 |
|
4 |
DCPS |
Decapping enzyme, scavenger |
2.341 |
-6.662 |
down |
1.28E-119 |
4 |
|
5 |
SMAD2 |
SMAD family member 2 |
1.412 |
-8.572 |
down |
1.16E-13 |
5 |
|
6 |
TRIM33 |
Tripartite motif containing 33 |
2.441 |
6.609 |
up |
6.99E-09 |
6 |
|
7 |
CHCHD2 |
Coiled-coil-helix-coiled-coil-helix domain containing 2 |
2.181 |
-5.569 |
down |
0.00E+00 |
7 |
|
8 |
NDUFA12 |
NADH:ubiquinone oxidoreductase subunit A12 |
1.766 |
-6.035 |
down |
2.04E-239 |
8 |
|
9 |
ATXN10 |
Spinocerebellar ataxia type 10 protein |
1.494 |
-6.501 |
down |
0.00E+00 |
9 |
|
10 |
CENPJ |
Centromere protein J |
1.681 |
7.13 |
up |
5.83E-11 |
10 |
|
11 |
MALT1 |
MALT1 paracaspase |
1.082 |
-9.366 |
down |
5.14E-58 |
11 |
|
12 |
LRCH1 |
Leucine rich repeats and calponin homology domain containing 1 |
2.253 |
-4.978 |
down |
4.06E-12 |
12 |
|
13 |
USP7 |
Ubiquitin specific peptidase 7 |
2.565 |
6.044 |
up |
5.45E-25 |
13 |
|
14 |
TELO2 |
Telomere maintenance 2 |
2.333 |
-4.878 |
down |
1.16E-79 |
14 |
|
15 |
NOTCH3 |
Notch receptor 3 |
1.825 |
6.635 |
up |
4.52E-04 |
15 |
|
16 |
MYBBP1A |
MYB binding protein 1a |
1.126 |
-7.007 |
down |
2.66E-120 |
16 |
|
17 |
RBM28 |
RNA binding motif protein 28 |
0.912 |
-10.38 |
down |
1.71E-85 |
17 |
|
18 |
TCTN3 |
Tectonic family member 3 |
1.113 |
-6.302 |
down |
2.98E-136 |
18 |
|
19 |
LARS1 |
Leucyl-tRNA synthetase 1 |
2.282 |
-4.471 |
down |
2.30E-08 |
19 |
|
20 |
TMEM216 |
Transmembrane protein 216 |
2.404 |
-4.387 |
down |
1.57E-18 |
20 |
|
21 |
PRMT1 |
Protein arginine methyltransferase 1 |
1.405 |
-4.939 |
down |
4.54E-30 |
21 |
|
22 |
TAB2 |
TGF-beta activated kinase 1 (MAP3K7) binding protein 2 |
0.867 |
8.956 |
up |
9.37E-18 |
22 |
|
23 |
EPAS1 |
Endothelial PAS domain protein 1 |
1.154 |
6.638 |
up |
3.94E-16 |
23 |
|
24 |
ZNF408 |
Zinc finger protein 408 |
1.149 |
-5.169 |
down |
2.00E-56 |
24 |
|
25 |
STUB1 |
STIP1 homology and U-box containing protein 1 |
5.134 |
-3.738 |
down |
0.00E+00 |
25 |
|
26 |
COG5 |
Component of oligomeric golgi complex 5 |
1.162 |
6.356 |
up |
2.09E-17 |
26 |
|
27 |
RIF1 |
Replication timing regulatory factor 1 |
0.746 |
25.494 |
up |
2.57E-42 |
27 |
|
28 |
IFT52 |
Intraflagellar transport 52 |
1.139 |
6.267 |
up |
1.18E-13 |
28 |
|
29 |
HBS1L |
HBS1 like translational GTPase |
0.796 |
-7.863 |
down |
1.08E-43 |
29 |
|
30 |
AUP1 |
AUP1 lipid droplet regulating VLDL assembly factor |
3.445 |
4.731 |
up |
6.28E-112 |
30 |
|
31 |
CC2D2A |
Coiled-coil and C2 domain containing 2A |
1.629 |
5.348 |
up |
1.84E-03 |
31 |
|
32 |
NFS1 |
NFS1 cysteine desulfurase |
1.362 |
-4.557 |
down |
1.56E-11 |
32 |
|
33 |
GPATCH8 |
G-patch domain containing 8 |
0.76 |
9.194 |
up |
2.04E-38 |
33 |
|
34 |
CLUAP1 |
Clusterin associated protein 1 |
1.358 |
5.496 |
up |
1.52E-05 |
34 |
|
35 |
QKI |
QKI, KH domain containing RNA binding |
0.912 |
6.989 |
up |
2.23E-11 |
35 |
|
36 |
RELA |
RELA proto-oncogene, NF-kB subunit |
1.465 |
-4.249 |
down |
3.24E-55 |
36 |
|
37 |
GMPPB |
GDP-mannose pyrophosphorylase B |
1.341 |
-4.415 |
down |
4.22E-198 |
37 |
|
38 |
MFF |
Mitochondrial fission factor |
0.872 |
-5.811 |
down |
2.44E-19 |
38 |
|
39 |
DYNC1LI1 |
Dynein cytoplasmic 1 light intermediate chain 1 |
1.883 |
-3.872 |
down |
1.30E-203 |
39 |
|
40 |
NDE1 |
nudE neurodevelopment protein 1 |
0.83 |
-6.026 |
down |
6.30E-238 |
40 |
|
41 |
CD81 |
CD81 molecule |
1.037 |
-4.857 |
down |
8.65E-53 |
41 |
|
42 |
FBXW7 |
F-box and WD repeat domain containing 7 |
1.78 |
-3.848 |
down |
7.36E-06 |
42 |
|
43 |
DYRK1A |
Dual specificity tyrosine phosphorylation regulated kinase 1A |
0.859 |
6.837 |
up |
2.02E-56 |
43 |
|
44 |
FANCD2 |
FA complementation group D2 |
0.995 |
6.104 |
up |
9.67E-19 |
44 |
|
45 |
NT5C3A |
5'-nucleotidase, cytosolic IIIA |
0.774 |
7.563 |
up |
1.28E-41 |
45 |
|
46 |
IKBKG |
Inhibitor of nuclear factor kappa B kinase regulatory subunit gamma |
15.098 |
4.174 |
up |
5.86E-08 |
46 |
|
47 |
SUFU |
SUFU negative regulator of hedgehog signaling |
1.139 |
-4.435 |
down |
2.73E-55 |
47 |
|
48 |
HSD17B10 |
Hydroxysteroid 17-beta dehydrogenase 10 |
1.404 |
-4.104 |
down |
0.00E+00 |
48 |
|
49 |
LARP7 |
La ribonucleoprotein 7, transcriptional regulator |
1.99 |
4.669 |
up |
1.66E-06 |
49 |
|
50 |
CHMP4B |
Charged multivesicular body protein 4B |
1.034 |
5.857 |
up |
0.00E+00 |
50 |
Negative regulations of SMAD2 lead to lung injury and upregulation of SMAD2 may trigger TGF-β signaling pathways that may lead to the downregulation of angiotensin-converting enzyme 2(ACE2) receptor and limit the spike protein interaction with ACE2 in the early phase. Other interesting genes from PBMC sample analysis were come out from our analysis are FBXW7, SMC4, CFL2, ATAD3A, ACAD8, RASSF1, SOX2, IGSF8, SP110, EHMT1, NODAL, PPAN, FAM118B, IFT172, SYNC, SH3GLB1, PTK2B, BIRC7, RUFY1, CLUAP1, PPT1, and AGAP3 that need to be investigated further for their role in SARS-CoV-2 infections.
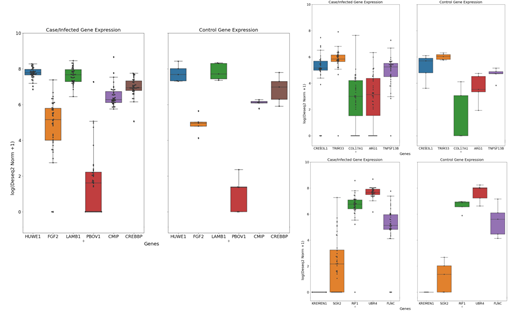
Lung tissues are the most important organ that gets affected during this disease. We identified TNF Superfamily Member 13B (TNFSF13B) as the most important transcription factor that is significantly differentially expressed in liver tissues with 2.955 Log2FC as a defense response suggesting its potential role in disease pathophysiological relations in SARS-CoV-2 infections [46]. It is supposed to activate B-cell and secretes antibodies into the blood and lung tissues that protect against pathogens and viral infection [47]. Whereas, Inducible T Cell Costimulator (ICOS) plays an important role in Tfh cell activation and high-affinity antibodies generation. In our study, we found it is significantly upregulated in disease conditions by 2.314 Log2FC and has multiple direct connections in the CoVint network. Various investigators have previously reported that downregulation of ICOS transcription factor significantly reduces Tfh cells [48–50] may lead to B cell differentiation and effective promotion of humoral immune responses [51] and helps in clearing life-threatening viruses including SARS-CoV-2. Therefore, potential inhibitors can be further explored for these proposed targets (Figure 10).
Lung tissues have also shown several genes that are significantly downregulated in disease conditions and especially in lung tissues. E3 ubiquitin-protein ligase (HUWE1) is one of them. It mediates ubiquitination and subsequent proteasomal degradation of target proteins [52–54]. Previously, the HUWE1 gene was reported to induce cell apoptosis in MERS-CoV ORF3 via ubiquitination. Activation of HUWE1 can play an antiviral role in host immunity [102] and can be further explored for potential therapeutic candidature. We have also identified another important transcriptional factor gene DDX17 that is significantly downregulated in our studied sample with a -15.742 Log2 (fold change). It was also scored as an important protein from our network analysis which represents this protein as a potential biomarker for SARS-CoV-2. DEAD-box (DDX) RNA helicases play a very crucial role in the stage of Covid-19 infection and therefore can be used as positive or negative regulators in different levels of DDX-mediated viral replication steps.
Table 5: The DEGs of merged datasets from the Lung sample with the applied criteria of p-value <0.05 and |log2FC| > 1.5, network score and directionality of regulations
|
Lung tissues |
|||||||
|
# |
Gene Symbol |
Gene Name |
Network Score |
Gene Expression |
Direction |
p-value |
Relative ranking |
|
1 |
TNFSF13B |
TNF superfamily member 13b |
2.902 |
2.955 |
up |
1.63E-07 |
1 |
|
2 |
ICOS |
Inducible T cell costimulator |
2.906 |
2.314 |
up |
9.20E-03 |
2 |
|
3 |
HUWE1 |
HECT, UBA and WWE domain containing E3 ubiquitin protein ligase 1 |
2.171 |
-15.978 |
down |
0.00E+00 |
3 |
|
4 |
LRRK2 |
Leucine rich repeat kinase 2 |
7.025 |
-14.834 |
down |
4.09E-141 |
4 |
|
5 |
CREBBP |
CREB binding protein |
2.121 |
-15.198 |
down |
7.35E-230 |
5 |
|
6 |
DDX17 |
DEAD-box helicase 17 |
1.414 |
-15.742 |
down |
0.00E+00 |
6 |
|
7 |
BAZ1B |
Bromodomain adjacent to zinc finger domain 1B |
2.709 |
-14.784 |
down |
7.80E-269 |
7 |
|
8 |
PLEKHA4 |
Pleckstrin homology domain containing A4 |
22.555 |
-14.334 |
down |
5.10E-125 |
8 |
|
9 |
EPAS1 |
Endothelial PAS domain protein 1 |
1.303 |
-15.763 |
down |
2.18E-117 |
9 |
|
10 |
MACF1 |
Microtubule actin crosslinking factor 1 |
1.123 |
-18.03 |
down |
3.37E-298 |
10 |
|
11 |
MDN1 |
Midasin AAA ATPase 1 |
1.387 |
-15.107 |
down |
2.95E-182 |
11 |
|
12 |
EP300 |
E1A binding protein p300 |
1.657 |
-14.678 |
down |
5.87E-306 |
12 |
|
13 |
UPF2 |
UPF2 regulator of nonsense mediated mRNA decay |
2.932 |
-14.233 |
down |
1.48E-220 |
13 |
|
14 |
UBQLN1 |
UBQLN1 |
3.81 |
-14.09 |
down |
1.15E-177 |
14 |
|
15 |
TAF15 |
TATA-box binding protein associated factor 15 |
1.302 |
-14.761 |
down |
1.02E-285 |
15 |
|
16 |
DOCK5 |
Dedicator of cytokinesis 5 |
2.375 |
-14.166 |
down |
1.41E-220 |
16 |
|
17 |
SMC3 |
Structural maintenance of chromosomes 3 |
3.775 |
-13.927 |
down |
1.40E-209 |
17 |
|
18 |
USP9X |
Ubiquitin specific peptidase 9 X-linked |
1.494 |
-14.351 |
down |
6.33E-282 |
18 |
|
19 |
WDR26 |
WD repeat domain 26 |
1.265 |
-14.726 |
down |
5.68E-262 |
19 |
|
20 |
SORT1 |
Sortilin 1 |
0.953 |
-15.359 |
down |
2.86E-278 |
20 |
|
21 |
HNRNPU |
Heterogeneous nuclear ribonucleoprotein U |
1.041 |
-15.208 |
down |
1.06E-285 |
21 |
|
22 |
AKAP9 |
A-kinase anchoring protein 9 |
0.861 |
-15.63 |
down |
1.50E-234 |
22 |
|
23 |
PIK3R1 |
Phosphoinositide-3-kinase regulatory subunit 1 |
0.835 |
-15.638 |
down |
5.33E-227 |
23 |
|
24 |
TRIM33 |
Tripartite motif containing 33 |
2.772 |
-13.791 |
down |
3.62E-178 |
24 |
|
25 |
SMAD2 |
SMAD family member 2 |
1.315 |
-14.264 |
down |
7.46E-90 |
25 |
|
26 |
SUPT16H |
SPT16 homolog, facilitates chromatin remodeling subunit |
7.35 |
-13.645 |
down |
2.29E-226 |
26 |
|
27 |
CHD7 |
Chromodomain helicase DNA binding protein 7 |
1.368 |
-14.129 |
down |
1.61E-145 |
27 |
|
28 |
HNRNPD |
Heterogeneous nuclear ribonucleoprotein D |
0.785 |
-15.452 |
down |
9.61E-276 |
28 |
|
29 |
YWHAE |
Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein epsilon |
0.786 |
-15.188 |
down |
1.73E-269 |
29 |
|
30 |
CDK12 |
Cyclin dependent kinase 12 |
0.889 |
-14.656 |
down |
4.66E-177 |
30 |
|
31 |
MAP1LC3B |
Microtubule associated protein 1 light chain 3 beta |
3.492 |
-13.499 |
down |
1.12E-236 |
31 |
|
32 |
CD81 |
CD81 molecule |
0.847 |
-14.656 |
down |
1.37E-273 |
32 |
|
33 |
PLEC |
Plectin |
0.638 |
-17.392 |
down |
1.12E-246 |
33 |
|
34 |
SAMHD1 |
SAM and HD domain containing deoxynucleoside triphosphate triphosphohydrolase 1 |
2.623 |
-13.501 |
down |
3.19E-92 |
34 |
|
35 |
RPL36 |
Ribosomal protein L36 |
1.392 |
-13.731 |
down |
5.16E-168 |
35 |
|
36 |
GRB2 |
Growth factor receptor bound protein 2 |
2.649 |
-13.447 |
down |
1.18E-197 |
36 |
|
37 |
ACTG1 |
Actin gamma 1 |
0.584 |
-17.213 |
down |
2.15E-258 |
37 |
|
38 |
KDM5B |
Lysine demethylase 5B |
1.643 |
-13.579 |
down |
9.26E-177 |
38 |
|
39 |
PTPN11 |
Protein tyrosine phosphatase non-receptor type 11 |
1.076 |
-13.895 |
down |
5.26E-303 |
39 |
|
40 |
UBC |
Ubiquitin C |
1.506 |
-13.536 |
down |
1.05E-94 |
40 |
|
41 |
AFG3L2 |
AFG3 like matrix AAA peptidase subunit 2 |
3.387 |
-13.224 |
down |
1.31E-141 |
41 |
|
42 |
ACTB |
Actin beta |
1.332 |
-13.624 |
down |
4.62E-109 |
42 |
|
43 |
SRPK1 |
SRSF protein kinase 1 |
1.181 |
-13.693 |
down |
1.47E-218 |
43 |
|
44 |
PRKDC |
Protein kinase, DNA-activated, catalytic subunit |
0.609 |
-15.145 |
down |
1.51E-297 |
44 |
|
45 |
BICD2 |
BICD cargo adaptor 2 |
1.929 |
-13.299 |
down |
4.50E-160 |
45 |
|
46 |
RAC1 |
Rac family small GTPase 1 |
0.752 |
-14.219 |
down |
8.96E-284 |
46 |
|
47 |
IQGAP1 |
IQ motif containing GTPase activating protein 1 |
0.575 |
-15.243 |
down |
0.00E+00 |
47 |
|
48 |
SBDS |
SBDS ribosome maturation factor |
2.001 |
-13.242 |
down |
4.17E-173 |
48 |
|
49 |
MYBBP1A |
MYB binding protein 1a |
1.15 |
-13.633 |
down |
1.80E-141 |
49 |
|
50 |
SUGT1 |
SGT1 homolog, MIS12 kinetochore complex assembly cochaperone |
1.993 |
-13.218 |
down |
6.72E-78 |
50 |
We also identified Endothelial PAS domain-containing protein 1 (EPAS1) also known as HIF2A is an important transcription factor that is involved in the regulation of oxygen level during the disease condition. However, inhibition of EPAS1 may not benefit in the early stage Covid-19 infection but may serve as a promising treatment target post-Covid-19 complication [56]. Another important protein Cyclic adenosine monophosphate response element-binding protein (CREBBP) found significantly downregulated in our study with a -15.198 Log 2-fold change value and serves as a potential biomarker for SARS-CoV-2. The functions of CREBBP are involved in cholesterol biosynthesis and Insulin regulation [57]. Dysregulation of cholesterol biosynthesis of Insulin could be an interesting MoA that several studies suggested its functional role that may lead to the reduction of SARS-CoV-2 infections [58] and regulating the entry of the SARS-CoV-2 virus into the host cell [58,59].
4. Conclusion
Our study has summarized the transcriptomics analysis of Covid-19 HTS data from Blood, PBMC and Lung tissues. We have identified 139 common DEGs from PBMC and blood, 291 common DEGs from Lung and Blood and 326 common DEGs from Lung and Blood. Our functional and CoVint network analysis revealed that the most common DEG from blood, PBMC and Lung tissue is CREB3L1, SOX2, UBR4 and FLNC. We also identified that ITPA, DLG3, ING4, TECR, NADH, SMAD, HUWE1, DDX17 and CREBBP are highly downregulated and activation of these genes may play an important role in disease reversal. Similarly, inhibition of RELA, HPSE, TRIM33, and TNFSF13B can be further explored experimentally for better therapeutic development and benefit of Covid-19 patients. In summary, these hub genes can be considered potential therapeutic biomarkers for the diagnosis of SARS-CoV-2.
Funding:
None
Conflict of interest:
The authors declare no conflict of interest, financial or otherwise.
Acknowledgements
Author of this paper would like to thank Dr. Gunjan Bhardwaj, CEO and Founder of Partex NV, and Gaurav Tripathi, Co-founder of Innoplexus Consulting Pvt Ltd, for their continuous support and motivation during this project. The author would also like to express gratitude to Komal Gowli and Aryamen Singh for their timely support in data sourcing and graph preparation. Additionally, sincere acknowledgments are extended to Mr. Holger Hoffman, Mr. Abhijit Keskar, and Mr. Amit Ananpara for their guidance, support, and provision of infrastructure facilities.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: COVID19 PBMC samples were retrieved from Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo) with accession number GSE166253 and GSE152418, accession number GSE150316. The whole blood RNA-seq dataset accession number is GSE185557, GSE166190, GSE169687, GSE161731 and GSE161777 and the COVID-19 lung RNA-seq dataset was retrieved from GSE151764 and GSE150316.
Author Contributions
O.S were responsible for the conception, design, and development of the methodology. RS was responsible for executing and identification of DEGs analysis. V.C was responsible for preparing CoVint internetwork. O.S performed, detailed analysis and validation of DEGs and overall scoring and hypothesis generation. O.S was responsible for the overall supervision of the study and writing the manuscript. All authors contributed to the article and approved the submitted version.
References
- Roshdy A, Zaher S, Fayed H & Coghlan JG. COVID-19 and the Heart: A Systematic Review of Cardiac Autopsies. Front Cardiovasc Med 7 (2020): 626975.
- Mokhtari T. et al. COVID-19 and multiorgan failure: A narrative review on potential mechanisms. J. Mol. Histol 51 (2020): 613–628.
- Hoffmann M. et al. The Omicron variant is highly resistant against antibody-mediated neutralization: Implications for control of the COVID-19 pandemic Cell 185 (2022): 447–456.e11.
- Hyland P. et al. Resistance to COVID-19 vaccination has increased in Ireland and the United Kingdom during the pandemic. Public Health 195 (2021): 54–56.
- Wang R, Chen J & Wei G-W. Mechanisms of SARS-CoV-2 Evolution Revealing Vaccine-Resistant Mutations in Europe and America. J. Phys. Chem. Lett 12 (2021): 11850–11857.
- CovMT: an interactive SARS-CoV-2 mutation tracker, with a focus on critical variants. Lancet Infect. Dis 21 (2021): 602.
- Kannan S, Shaik Syed Ali P & Sheeza A. Omicron (B.1.1.529) - variant of concern - molecular profile and epidemiology: a mini review. Eur. Rev. Med. Pharmacol. Sci 25 (2021): 8019–8022.
- The Lancet Infectious Diseases. Unmet need for COVID-19 therapies in community settings. Lancet Infect. Dis 21 (2021): 1471.
- di Iulio J, Bartha I, Spreafico R, Virgin HW & Telenti A. Transfer transcriptomic signatures for infectious diseases. Proc. Natl. Acad. Sci. U. S. A 118 (2021).
- Barrett T. et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res 41 (2013): D991–5.
- FelixKrueger. GitHub - FelixKrueger/TrimGalore: A wrapper around Cutadapt and FastQC to consistently apply adapter and quality trimming to FastQ files, with extra functionality for RRBS data.
- Patro R, Duggal G, Love MI, Irizarry RA & Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14 (2017): 417–419.
- Love MI, Huber W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15 (2014): 550.
- Sharma OP, Seiz W & Scheele J. SARS-CoV-2 therapeutic landscape, opportunity and future threats. The Open COVID Journal 1 (2021): 205–215.
- Mallick, I. et al. In-silico identification and prioritization of therapeutic targets of asthma. Sci. Rep 13 (2023): 15706.
- Ontosight® Explore. https://www.innoplexus.com/life-science-ai-products-solutions/ontosight-xplore/ (2018).
- United States Patent Application: 0200090789.
- Ostaszewski M. et al. COVID19 Disease Map, a computational knowledge repository of virus-host interaction mechanisms. Mol. Syst. Biol 17 (2021): e10387.
- Ashburner M. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet 25 (2000): 25–29.
- Ritchie ME. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43 (2015): e47.
- Luo J. et al. The potential involvement of JAK-STAT signaling pathway in the COVID-19 infection assisted by ACE2. Gene 768 (2021): 145325.
- Farahani M. et al. Molecular pathways involved in COVID-19 and potential pathway-based therapeutic targets. Biomed. Pharmacother 145 (2022): 112420.
- Mohanta TK, Sharma N, Arina P & Defilippi P. Molecular Insights into the MAPK Cascade during Viral Infection: Potential Crosstalk between HCQ and HCQ Analogues. Biomed Res. Int (2020): 8827752.
- Khalil BA, Elemam NM & Maghazachi AA. Chemokines and chemokine receptors during COVID-19 infection. Comput. Struct. Biotechnol. J 19 (2021): 976–988.
- Giacomelli C, Piccarducci R, Marchetti L, Romei C & Martini C. Pulmonary fibrosis from molecular mechanisms to therapeutic interventions: lessons from post-COVID-19 patients. Biochem. Pharmacol 193 (2021): 114812.
- Kiener M. et al. Human-Based Advanced Approaches to Investigate Lung Fibrosis and Pulmonary Effects of COVID-19. Front. Med 8 (2021): 644678.
- Greenwood M, Greenwood MP, Paton JFR & Murphy D. Transcription Factor CREB3L1 Regulates Endoplasmic Reticulum Stress Response Genes in the Osmotically Challenged Rat Hypothalamus. PLoS One 10 (2015): e0124956.
- Sajuthi SP, et al. Type 2 and interferon inflammation strongly regulate SARS-CoV-2 related gene expression in the airway epithelium. bioRxiv (2020).
- Zhang Z. et al. Clinical analysis and pluripotent stem cells-based model reveal possible impacts of ACE2 and lung progenitor cells on infants vulnerable to COVID-19. Theranostics 11 (2021): 2170–2181.
- Tiwari SK, Wang S, Smith D, Carlin AF & Rana TM. Revealing Tissue-Specific SARS-CoV-2 Infection and Host Responses using Human Stem Cell-Derived Lung and Cerebral Organoids. Stem Cell Reports 16 (2021): 437–445.
- Sano E. et al. Modeling SARS-CoV-2 infection and its individual differences with ACE2-expressing human iPS cells. iScience 24 (2021): 102428.
- Lee C-J. et al. Crosstalk between SOX2 and cytokine signaling in endometrial carcinoma. Sci Rep 8 (2018): 17550.
- Li S-W. et al. Severe acute respiratory syndrome coronavirus papain-like protease suppressed alpha interferon-induced responses through downregulation of extracellular signal-regulated kinase 1-mediated signalling pathways. J. Gen. Virol 92 (2011): 1127–1140.
- Page C. et al. Induction of alternatively activated macrophages enhances pathogenesis during severe acute respiratory syndrome coronavirus infection. J. Virol 86 (2012): 13334–13349.
- Almasy KM, Davies JP & Plate L. Comparative host interactomes of the SARS-CoV-2 nonstructural protein 3 and human coronavirus homologs. bioRxiv (2021).
- Tripathi S. et al. Meta- and Orthogonal Integration of Influenza ‘OMICs’ Data Defines a Role for UBR4 in Virus Budding. Cell Host Microbe 18 (2015): 723–735.
- Al-Eitan LN, & Alahmad SZ. Pharmacogenomics of genetic polymorphism within the genes responsible for SARS-CoV-2 susceptibility and the drug-metabolising genes used in treatment. Rev. Med Virol 31 (2021): e2194.
- Caillet-Saguy C. et al. Host PDZ-containing proteins targeted by SARS-CoV-2. FEBS J 288 (2021): 5148–5162.
- Yin X. et al. MDA5 Governs the Innate Immune Response to SARS-CoV-2 in Lung Epithelial Cells. Cell Rep 34 (2021): 108628.
- Shi C. et al. The Potential of Low Molecular Weight Heparin to Mitigate Cytokine Storm in Severe COVID-19 Patients: A Retrospective Cohort Study. Clin. Transl. Sci 13 (2020): 1087–1095.
- Kiyan Y. et al. Heparanase-2 protects from LPS-mediated endothelial injury by inhibiting TLR4 signalling. Sci. Rep 9 (2019): 13591.
- Ali H, et al. Cellular TRIM33 restrains HIV-1 infection by targeting viral integrase for proteasomal degradation. Nat. Commun 10 (2019): 926.
- Martonik D, Parfieniuk-Kowerda A, Rogalska M, & Flisiak R. The Role of Th17 Response in COVID-19 Cells 10 (2021).
- Tanaka S. et al. Trim33 mediates the proinflammatory function of Th17 cells. J Exp Med 215 (2018): 1853–1868.
- Vaz de Paula CB. et al. COVID-19: Immunohistochemical Analysis of TGF-β Signaling Pathways in Pulmonary Fibrosis. Int J Mol Sci 23 (2021).
- Gelarden I. et al. Comprehensive evaluation of bronchoalveolar lavage from patients with severe COVID-19 and correlation with clinical outcomes. Hum. Pathol 113 (2021): 92–103.
- Sosa-Hernández VA, et al. B Cell Subsets as Severity-Associated Signatures in COVID-19 Patients. Front. Immunol 11 (2020): 611004.
- Cui D. et al. Follicular Helper T Cells in the Immunopathogenesis of SARS-CoV-2 Infection. Front. Immunol 12 (2021): 731100.
- Hutloff A. Regulation of T follicular helper cells by ICOS. Oncotarget 6 (2015): 21785–21786.
- Huang X. et al. The ubiquitin ligase Peli1 inhibits ICOS and thereby Tfh-mediated immunity. Cell. Mol. Immunol 18 (2021): 969–978.
- Weber JP, et al. ICOS maintains the T follicular helper cell phenotype by down-regulating Krüppel-like factor 2. J. Exp Med 212 (2015): 217–233.
- Zhong Q, Gao W, Du F & Wang X. Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell 121 (2005): 1085–1095.
- Parsons JL, et al. Ubiquitin ligase ARF-BP1/Mule modulates base excision repair. EMBO J 28 (2009): 3207–3215.
- Yoon SY. et al. Over-expression of human UREB1 in colorectal cancer: HECT domain of human UREB1 inhibits the activity of tumor suppressor p53 protein. Biochem. Biophys. Res. Commun 326 (2005): 7–17.
- Zhou Y, et al. Host E3 ligase HUWE1 attenuates the proapoptotic activity of the MERS-CoV accessory protein ORF3 by promoting its ubiquitin-dependent degradation. J. Biol Chem 298 (2022): 101584.
- Poloznikov AA, et al. HIF Prolyl Hydroxylase Inhibitors for COVID-19 Treatment: Pros and Cons. Front. Pharmacol 11 (2020): 621054.
- Zhou XY. et al. Insulin regulation of hepatic gluconeogenesis through phosphorylation of CREB-binding protein. Nat. Med 10 (2004): 633–637.
- Ziegler CGK, et al. Impaired local intrinsic immunity to SARS-CoV-2 infection in severe COVID-19. bioRxiv (2021).
- Kocar E, Rezen T & Rozman D. Cholesterol, lipoproteins, and COVID-19: Basic concepts and clinical applications. Biochim. Biophys. Acta Mol. Cell Biol Lipids 1866 (2021): 158849.


 Impact Factor: * 6.2
Impact Factor: * 6.2 Acceptance Rate: 76.33%
Acceptance Rate: 76.33%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 