Analysis of the Gut Microbiota in Suncus murinus, a Natural Obesity- Resistant Experimental Animal
Mingshou Zhang1, Ting Yang1, Rujia Li1, Shunichi Uetake1, Ke Ren2, Jun Li3, Shuang-Qin Yi1*
1Department of Frontier Health Sciences, Graduate School of Human Health Sciences, Tokyo Metropolitan University, 116-8551 Tokyo, Japan
2Faculty of Physical Education, Qu Jing Normal University, Yun Nan, China
3State Key Laboratory of Oncogenes and Related Genes, Shanghai Cancer Institute, Ren Ji Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai, China
*Corresponding Author: Shuang-Qin Yi, Department of Frontier Health Sciences, Graduate School of Human Health Sciences, Tokyo Metropolitan University, 7-2-10, Higashiogu, Arakawa-ku, 116-8551 Tokyo, Japan.
Received: 16 November 2023; Accepted: 23 November 2023; Published: 22 February 2024
Article Information
Citation: Mingshou Zhang, Ting Yang, Rujia Li, Shunichi Uetake, Ke Ren, Jun Li, Shuang-Qin Yi. Analysis of the Gut Microbiota in Suncus murinus, a Natural Obesity-Resistant Experimental Animal. Journal of Food Science and Nutrition Research. 7 (2024): 44-51.
DOI: 10.26502/jfsnr.2642-110000151
View / Download Pdf Share at FacebookAbstract
In our recent studies, we investigated the phenomenon of obesity resistance in the Asian house shrew, Suncus murinus, which may be a suitable model to study the mechanisms of obesity resistance. In this study, we characterized the gut microbiota of S. murinus by analyzing the microbiota using libraries of cloned bacterial 16S rRNA gene sequences to explore their relationship with natural obesity-resistance properties. Our findings revealed a distinct microbial profile of S. murinus, primarily dominated by Firmicutes and Proteobacteria, with a notable absence of Bacteroidetes. This composition is consistent with the obesity-resistance properties of animals. The low microbiota diversity observed in S. murinus may be associated with its unique gastrointestinal tract morphology, lack of fermentative chambers (such as a cecum), and insectivorous dietary habits. Lactic acid bacteria were abundant in the gut microbiota of S. murinus, suggesting the potential importance of lactic acid fermentation processes in this species. Helicobacter, a known human pathogen, is also present in significant quantities in the gut of S. murinus. Further research is warranted to explore the specific functions and interactions of these microbial groups in S. murinus and their broader ecological implications to contribute to the study of the mechanism of obesity in humans.
Keywords
<p>Obesity, Gut Microbiota, Lactic acid bacteria</p>
Article Details
Introduction
The Asian house shrew, Suncus murinus (S. murinus), is an insectivorous animal that commonly inhabits houses and grassy areas near human dwellings or livestock enclosures. Its distribution spans a broad geographical range, encompassing Southeast Asia and East Africa [1,2]. S. murinus belongs to the Eulipotyphla phylum, a part of the ancient superorder Laurasiatheria, which dates back to a time over 66 million years ago, well before the Cretaceous-Paleogene boundary. It diverged from the superorder Euarchontoglires, encompassing humans, mice, rats, and rabbits, long before this boundary [3]. Japan exhibits native populations of this species on the Nansei Islands, such as the Amami, Okinawa, Miyako, and Yaeyama Islands, while introduced populations have successfully colonized Kyushu [4].
Members of the phylum Eulipotyphla display a multitude of primitive mammalian morphological characteristics. They primarily feed on insects and have a simple digestive system without specialized compartments like the cecum and forestomach. In terms of histology, their large intestine extends a mere 1 cm from the anus [3]. S. murinus exhibits distinct features in its gastrointestinal tract, characterized by a straightforward structure without fermentation chambers, a relatively short overall intestine length, and an exceptionally abbreviated large intestine [5]. The distinct traits observed in S. murinus can be largely attributed to their evolutionary history.
Since the 1970s, this species has been successfully domesticated as a valuable laboratory resource, leading to the development of multiple strains from diverse geographical sources, all designed to facilitate research efforts [6,7]. S. murinus has been used in various fields of science because of its unique characteristics as a model for studying emesis [8], conducting morphological analyses of nerves and the gastrointestinal tract, developing pathological models [9,10], and evaluating in vivo motilin functions [11].
One intriguing aspect of S. murinus is its distinctive resistance to obesity, a characteristic that sets it apart from other laboratory rodents. In our recent study, we investigated the obesity resistance phenomenon in S. murinus. It was found that the weight of S. murinus did not change much after two months old, and mesenteric fat did not accumulate after birth [12,13]. Thus, S. murinus may be a suitable model for investigating obesity and metabolic syndrome, particularly the mechanisms of obesity resistance [12]. However, the mechanism underlying natural obesity resistance is still not well understood. To continue research on this natural obesity-resistant animal, we focused on the microbes that inhabit its intestines.
In the present study, we characterized the gut microbiota of S. murinus by analyzing the microbiota using libraries of cloned bacterial 16S rRNA gene sequences to explore their relationship with natural obesity-resistance properties.
Materials and Methods
Animals
Male S. murinus animals (n=5; age, 8 weeks old) were obtained from a closed breeding colony (JIc: KAT-c, at our laboratory) [13,14]. All animals were housed in polycarbonate cages in a room maintained at 28 ± 2 °C with 50% ± 5% relative humidity at the Functional Morphology Laboratory in the Department of Frontier Health Sciences, Tokyo Metropolitan University (Tokyo, Japan). The room was automatically lit between 09:00 and 21:00. The pellets consisted of 45.0% protein, 4.0% fat, 3.0% fiber, 15.0% ash, and 26.2% complex carbohydrates (Bioindustry Division of Oriental Yeast Co., Ltd., Chiba, Japan). The metabolizable energy content was 357 kcal/100 g, and the pellets and water were supplied ad libitum.
Animal Experimentation Arrival Guidance Statement
This study was reported in accordance with ARRIVE guidelines ([https://arriveguidelines.org)] https://arriveguidelines.org).
All animal experiments were approved by the Institutional Animal Care and Use Committee of Tokyo Metropolitan University (permit number: A2-2, A3-22, A4-19, A5-14). All experimental procedures were performed according to the “National Research Council Guide for Care and Use of Laboratory Animals”.
After the experiment was completed, the animals were sacrificed by inhaling an overdose of the anesthetic isoflurane (concentration 5.0%, v/v) using a continuous inhalation anesthesia machine (SN-487-OT Air, WAKENYAKU CO., LTD, Kyoto, Japan).
Fecal microbiota collection and extraction of DNA from feces
To collect fecal content from S. murinus, first, all surgical instruments were aseptically sterilized for sample collection. S. murinus were deeply anesthetized under inhalation anesthesia with isoflurane (concentration 2.5%, v/v) using a continuous inhalation anesthesia machine (SN-487-OT Air, WAKENYAKU CO.,LTD, Kyoto, Japan), and then the abdominal skin was disinfected with iodine, and a midline abdominal incision was made. The large intestine (S. murinus has no cecum) was cut open, and approximately 200 mg of content was collected and transferred into a 2 mL sterile test tube, with care taken to avoid external contamination. After feces collection, experimental animals were euthanized by overdose of anesthesia.
The collected feces were immediately dissolved with the reagents in the DNA Kit, and fecal DNA was extracted from samples using an ISOSPIN Fecal DNA Kit (Nippon Gene Co., Ltd., Toyama, Japan) according to the manufacturer’s protocols. A NanoDrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DF, USA) was used to determine the concentration of extracted DNA. Samples that did not meet the detection standards were removed. All DNA samples were stored at –80 °C immediately before the next process.
Library construction and sequencing
The library construction and sequencing part of this study was commissioned by a professional and authoritative company (Pharma Foods International Co. Ltd., Tokushima, Japan), APRO Science Group (https://apro-s.com/service-index/) to complete the analysis. In brief, sequencing libraries were prepared according to the Illumina 16S Metagenomic Sequencing Library protocols to amplify the V3–V4 region. The input gDNA 2 ng was amplified via polymerase chain reaction (PCR) with 5× reaction buffer, 1 mM dNTP mix, 500 nM each of the universal F/R PCR primers, and Herculase II fusion DNA polymerase (Agilent Technologies, Santa Clara, CA, USA). The first PCR cycle conditions were as follows: 3 min at 95 °C for heat activation, 25 cycles of 30 s at 95 °C, 30 s at 55 °C, and 30 s at 72 °C, followed by a 5-min final extension at 72 °C. The universal primer pair with Illumina adapter overhang sequences used for the first amplification was as follows: V3-F:5’-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG-3,’ V4-R:5’- GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC-3’. The first PCR product was purified using AMPure beads (Agencourt Bioscience, Beverly, MA, USA). Following purification, 2 μl of the first PCR product was PCR-amplified for the final library construction index using the NexteraXT Indexed Primer. The second PCR cycle conditions were the same as the first conditions, except that there were 10 cycles. The PCR products were purified using AMPure beads. The final purified product was then quantified by qPCR according to the qPCR Quantification Protocol Guide (KAPA Library Quantification kits for Illumina Sequencing platforms) and qualified using TapeStation D1000 ScreenTape (Agilent Technologies, Waldbronn, Germany). Paired-end (2×300 bp) sequencing was performed by Macrogen (Soul, South Korea) using the MiSeq platform (Illumina, San Diego, CA, USA).
Sequence analyses
For adapter trimming, adapter sequences were removed, and error correction was performed for areas where the two reads overlapped using fastp (v. 0.19.7) [15]. Subsequently, the original library and single long reads were obtained by assembling paired-end sequences created by sequencing both directions of the library using the FLASH software program (v.1.2.11) [16].
Operational taxonomic units (OTUs) were clustered with a cutoff value set at 97% similarity, and chimeric sequences were identified and removed using CD-HIT-OTU (cd-hit-otu-illumina-0.0.1, http://weizhong-lab.ucsd.edu/cd-hit-otu) [17]. A BLAST+(v2.9.0) search (query coverage >85% and identity >85%) was performed for each 16S rRNA gene sequence against the RDP Release 11 database (RDP Release 11 Update 4: May 26) to obtain taxonomic information.
Bioinformatics and statistical analyses
QIIME (V.1.9.1) [18] and the R packages (version 3.4.4, https://www.r-project. org/) were used for the sequencing analysis of the gut microbiota. Statistical differences in alpha diversity indices (i.e. the Shannon index, inverse Simpson index, observed richness, Chao’s estimated richness [Chao1], and relative abundance of the different taxonomic groups) were measured using the Kruskal-Wallis test and Wilcoxon’s t-test. Furthermore, bacterial community diversity was analyzed using rarefaction plots and displayed using the R software program (version 3.4.4).
T-tests were used for the statistical analysis using R (version 3.4.4), and results were presented as the mean with the standard error of mean (SEM). P values of <0.05 were considered statistically significant.
Results
Overview of the sequencing data
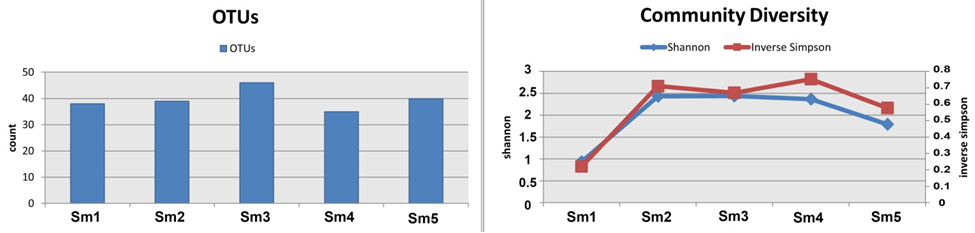
High-quality reads corresponding to OTUs were identified after filtering chimeric and low-quality OTUs. Good coverage was up to 99.9%, while only 35-46 OTUs were detected in S. murinus. The alpha diversity (Shannon, Chao1, and Inverse Simpson) of the microbial communities was assessed and calculated. No significant differences were observed among the samples (Table 1 and Figure 1).
|
Sample |
OTUs |
Chao1 |
Shannon |
Inverse Simpson |
Good's Coverage |
|
Sm1 |
38 |
39 |
0.93953229 |
0.2216564 |
0.999886265 |
|
Sm2 |
39 |
39 |
2.434212501 |
0.710451015 |
0.999944626 |
|
Sm3 |
46 |
50 |
2.438446446 |
0.669201789 |
0.999801351 |
|
Sm4 |
35 |
42 |
2.369102485 |
0.753479491 |
0.999723626 |
|
Sm5 |
40 |
40.25 |
1.790979773 |
0.577638118 |
0.999941958 |
OTUs, operational taxonomic unit; Chao1, returns the Chao1 richness estimate for an OTU definition; Shannon, the Shannon index takes into account the number and evenness of species; Inverse Simpson, the Inverse Simpson index represents the probability that two randomly selected individuals in the habitat will belong to the same species; Good's Coverage, Coverage is calculated as C=1-(s/n).
Table 1: Diversity and richness (mean ± SD) of the fecal bacteria communities of mice and S. murinus
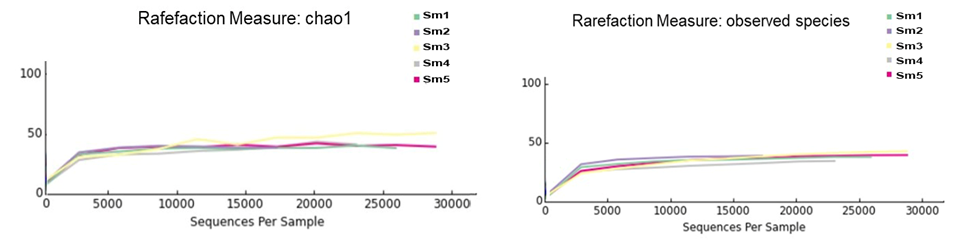
The rarefaction curves (Figure 2) showed the number of species/OTUs under different sequence numbers when the rarefaction curves approached a plateau, indicating that the number of reads used in the analysis and the alpha diversity of the sampled community were sufficiently extrapolated (Figure 2). These data show that the results are reasonable and reliable for the subsequent analyses.
Bacteria composition and relative abundance
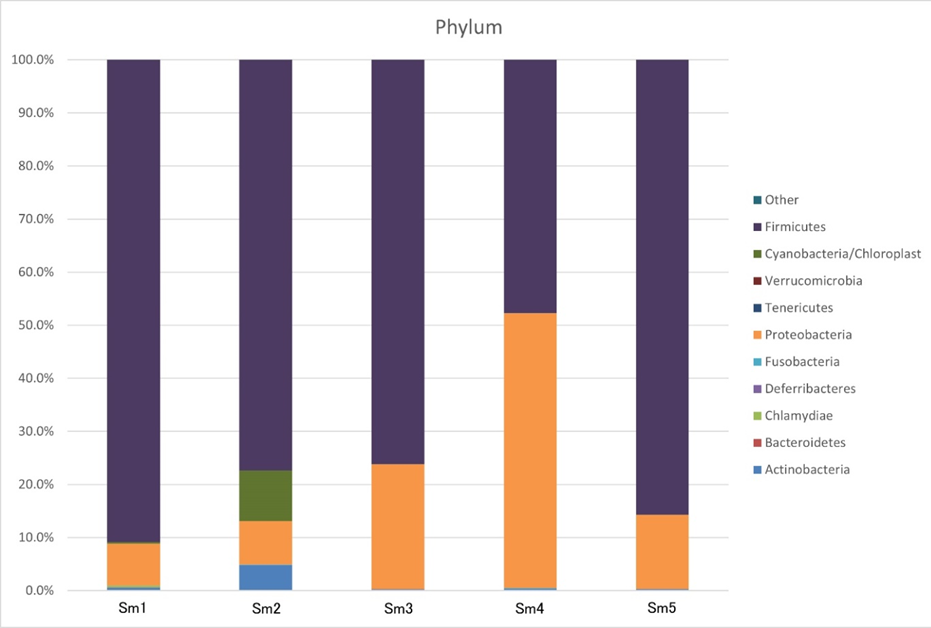
We chose the main phyla and genera based on species abundance to generate a histogram showing the percentages of relative abundance in each sample (Figure 3).
At the phylum level (Figure 3, Table 2), the most abundant phyla were Firmicutes (47.71%-90.86%) and Proteobacteria (7.87%-51.78%) in each sample, and only 0.26%-4.85% of Actinobacteria were detected in each sample; we did not detect any sequences identified as Bacteroidetes (0.00%), Deferribacteres (0.00%), Tenericutes (0.00%), or Verrucomicrobia (0.00%) in any sample, even though these are major bacterial phyla in the gastrointestinal tracts of humans and mice. Other phyla (Chlamydiae, Fusobacteria, and Cyanobacteria/Chloroplast) accounted for <1%.
Sm 1-5, samples of S. murinus
Table 2: Each sample composition percentage at phylum level.
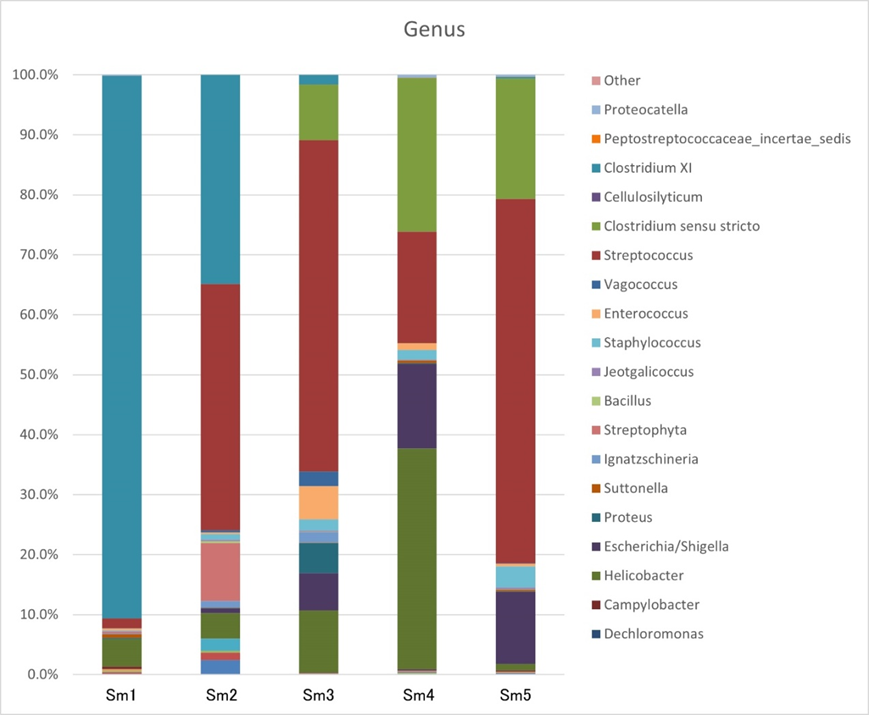
At the genus level (Figure 4, Table 3), the most abundant genera were Streptococcus (35.27%±24.91%), Clostridium XI (24.92%±16.87%), Helicobacter (11.35%±14.53%), Clostridium sensu stricto (10.96%±8.29%), Escherichia/Shigella (6.62%±5.98), Streptophyta (1.97%±4.72), Staphylococcus (1.61%±1.27%), Enterococcus (1.52%±2.26%), and Vagococcus (0.59%±1.04%). Most belonged to Lactobacillales. Other bacterial genera accounted for <0.05%.
|
Genus |
Sm1 |
Sm2 |
Sm3 |
Sm4 |
Sm5 |
|
Corynebacterium |
0.13% |
2.38% |
0.18% |
0.13% |
0.28% |
|
Okibacterium |
0.24% |
1.26% |
0.00% |
0.00% |
0.00% |
|
Euzebya |
0.22% |
0.28% |
0.07% |
0.23% |
0.02% |
|
Leptotrichia |
0.00% |
0.03% |
0.00% |
0.15% |
0.01% |
|
Bradyrhizobium |
0.06% |
1.97% |
0.00% |
0.00% |
0.00% |
|
Brachymonas |
0.25% |
0.00% |
0.02% |
0.15% |
0.18% |
|
Dechloromonas |
0.07% |
0.00% |
0.00% |
0.14% |
0.02% |
|
Campylobacter |
0.38% |
0.00% |
0.02% |
0.12% |
0.19% |
|
Helicobacter |
4.47% |
4.19% |
10.35% |
36.66% |
1.10% |
|
Escherichia/Shigella |
0.00% |
0.79% |
6.23% |
14.06% |
12.00% |
|
Proteus |
0.16% |
0.08% |
5.00% |
0.14% |
0.07% |
|
Suttonella |
0.63% |
0.06% |
0.06% |
0.42% |
0.25% |
|
Ignatzschineria |
0.11% |
1.06% |
1.76% |
0.00% |
0.00% |
|
Streptophyta |
0.30% |
9.54% |
0.02% |
0.00% |
0.01% |
|
Bacillus |
0.00% |
0.28% |
0.02% |
0.00% |
0.06% |
|
Jeotgalicoccus |
0.10% |
0.23% |
0.19% |
0.11% |
0.32% |
|
Staphylococcus |
0.08% |
0.95% |
1.91% |
1.65% |
3.51% |
|
Enterococcus |
0.31% |
0.22% |
5.52% |
1.10% |
0.45% |
|
Vagococcus |
0.09% |
0.40% |
2.45% |
0.02% |
0.03% |
|
Streptococcus |
1.57% |
40.45% |
55.18% |
18.52% |
60.64% |
|
Clostridium sensu stricto |
0.00% |
0.00% |
9.23% |
25.52% |
20.05% |
|
Cellulosilyticum |
0.03% |
0.00% |
0.02% |
0.00% |
0.00% |
|
Clostridium XI |
88.48% |
34.33% |
1.59% |
0.00% |
0.24% |
|
Peptostreptococcaceae_incertae_sedis |
0.01% |
0.00% |
0.00% |
0.02% |
0.01% |
|
Proteocatella |
0.16% |
0.00% |
0.00% |
0.48% |
0.30% |
|
Other |
0.00% |
0.00% |
0.00% |
0.00% |
0.00% |
Sm 1-5, samples of S. murinus
Table 3: Each sample composition percentage at the genus level.
Discussion
In this study, 16S rRNA amplicon sequencing was used to comprehensively characterize the gut microbiota of S. murinus. The microbiota of S. murinus was enriched in Firmicutes (75.55%) and Proteobacteria (21.07%), whereas Bacteroidetes were not detected, which is consistent with a previous study [3]. S. murinus displays an unusual enrichment of the Proteobacteria phylum, a feature uncommon in rodents. Proteobacteria, which was also prominently present in the giant panda, is associated with lignin digestion and the breakdown of various components [19,20]. We hypothesized that the observed relative proportions of Firmicutes and Proteobacteria in our study could be linked to the obesity-resistant characteristics of this species.
murinus exhibits a lower gut microbiota diversity compared to other mammalian species. While conclusive evidence is lacking, there is speculation that this reduced diversity may be associated with the morphological characteristics of its gastrointestinal tract [3]. In our earlier research studies [5,11], it was shown that S. murinus does not possess fermentative chambers like the forestomach and cecum. These specific features may restrict the available physiological and physical niches for microbial communities. For instance, certain bacteria such as Bacteroidetes, including Bacteroidaceae, Prevotellaceae, and Rikenellaceae, are typically prevalent in the cecum [21]. Given the absence of a cecum in S. murinus, these particular bacteria were not identified in the gut during our current study.
murinus are insectivores, demonstrating food habits that differ from those of herbivores and omnivores. Therefore, we speculate that the diversity of the gut microbiota in S. murinus is not only related to the evolutionary background but also has a strong relationship with the diet structure. Ley et al. revealed that the diversity of the fecal microbiota in mammals is higher in herbivores than in omnivores and is lowest in carnivores [22]. This implies that food habits are an important factor influencing the gut microbiota.
In this current study, the gut of S. murinus exhibited a notable abundance of lactic acid bacteria at the genus level, including Enterococcus, Lactococcus, Streptococcus, and Vagococcus. This suggests that lactic acid fermentation could hold significance within the gastrointestinal ecosystem of S. murinus. This finding strongly indicates the potential importance of lactic acid fermentation processes in this species. Lactic acid bacteria have been shown to potentially impact factors related to obesity, such as metabolism, appetite regulation, immune system function, and the balance of intestinal flora, as demonstrated in human studies [23], lactic acid bacteria contribute to the complexity of gut microbiota and exert many beneficial effects on metabolism and immunomodulation.
Enterococcus is a genus of bacteria that falls under the category of lactic acid bacteria. Similar to other lactic acid bacteria. Enterococcus strains have probiotic properties and can be used in probiotic formulations to support gut health and improve digestion [24]. Lactococcus belongs to the lactic acid bacteria group. Through lactic acid production, Lactococcus helps preserve dairy products by creating an acidic environment that inhibits the growth of harmful bacteria and spoilage microorganisms [25]. Streptococcus is a genus of Gram-positive bacteria that includes a wide range of species. Vagococcus belongs to the family Enterococcaceae and is primarily present in the genital tract of animals, including humans [26]. Interestingly, Helicobacter spp. are abundant in the gut of S. murinus. Helicobacter species are significant human pathogens and are strongly associated with several gastric diseases. In addition to humans, Helicobacter species have been found in the stomachs of various animals, including primates, cats, dogs, and rodents. Some Helicobacter species have been studied for their potential impact on animal health [27]. It should be noted that these lactic acid bacteria and other members of Firmicutes are not core members of the human or mouse gut microbiota. These special intestinal flora structures may allow S. murinus to have natural obesity-resistant properties.
In summary, the present study focused on investigating the potential functions of the gut microbiota of S. murinus, an insectivorous animal. Through 16S rRNA-amplicon sequencing, the gut microbiota composition of S. murinus was thoroughly analyzed. The results revealed a unique microbial profile in S. murinus, characterized by a predominance of Firmicutes and Proteobacteria, with the absence of Bacteroidetes, which is consistent with its obesity-resistant properties. This difference in microbiota composition might be related to S. murinus' distinctive morphological features, such as the lack of fermentative chambers, like the cecum, and its food habits.
Overall, this study sheds light on the unique composition of the gut microbiota of S. murinus. However, more in-depth investigations are needed to fully comprehend the role and interactions of these specific bacterial groups in S. murinus and their broader ecological significance, especially concerning the natural obesity-resistant properties of this animal.
Author contributions
SY acquired the funding and designed and conceived the study. MZ and TY performed experiments. MZ, SY, and RL analyzed the data. MZ wrote the article. All authors have contributed to the final version of this manuscript. All authors have read and approved the final manuscript.
Data availability statement
The datasets generated and/or analysed during the current study are available from the GenBank of National Center for Biotechnology Information (NCBI) official website (accession number: OQ119157-OQ119540).
Conflicts of interest
The authors declare that they have no conflicts of interest.
References
- Yi SQ, Shimokawa T, Akita K, et al. Anatomical Study of the Pancreas in the House Musk Shrew (Suncus murinus), with Special Reference to the Blood Supply and Innervation. The Anatomical Record 273A (2003): 630-635.
- Yi SQ, Ohta T, Miwa K, et al. Surgical anatomy of the innervation of the major duodenal papilla in human and Suncus murinus, from the perspective of preserving innervation in organ-saving procedures. Pancreas 30 (2005): 211-217.
- Shinohara A, Nohara M, Kondo Y, et al. Comparison of the gut microbiotas of laboratory and wild asian house shrews (Suncus murinus) based on cloned 16s rRNA sequences. Experimental Animals 68 (2019): 531-539.
- Motokawa M. Suncus murinus (Linnaeus, 1766). In: the Wild Mammals of Japan, Second edition (Ohdachi SD, Ishibashi Y, Iwasa M, Fukui D, and Saitoh T. eds.), Shouladoh, Kyoto, Japan (2015): 26-27.
- Yi SQ, Li J, Yamaguchi T, et al. Immunolocalization of the PP family and its receptors in the gastrointestinal tract of house musk shrew, Suncus murinus. Neuroendocrinology Letters 32 (2011): 212-219.
- Oda S, Kondo K. Progress in domestication of Suncus murinus riukiuanus for experiment (in Japanese). The Mammal Society of Japan 33 (1976): 13-30.
- Oda S, Kondo K. Usefulness of wild insectivores as laboratory animals (in Japanese). Experimental Animals 26 (1977): 273-280.
- Ebukuro S, Wakana S, Hioki K, et al. Selective breeding of house musk shrew (Suncus murinus) lines in relation to emesis induced by veratrine sulfate. Comparative Medicine 50 (2000): 281-283.
- Tsutsui C, Kajihara K, Yanaka T, et al. House musk shrew (Suncus murinus, order: Insectivora) as a new model animal for motilin study. Peptides 30 (2009): 318-329.
- Yi SQ, Li J, Qu N, et al. House musk shrew, Suncus murinus: A novel and natural obesity-resistant animal model. Obesity & Metabolism 6 (2010): 22-28.
- Zhang M, Dai Y, Sasaki H, et al. High fat diet load study in a natural obesity-resistant animal model, Suncus murinus. Journal of Veterinary Medical Science 2 (2020): 19-29.
- Yi SQ, Ru F, Ohta T, et al. Surgical anatomy of the innervation of pylorus in human and Suncus murinus, in relation to surgical technique for pylorus-preserving pancreaticoduodenectomy. World Journal of Gastroenterology 12 (2006): 2209-2216.
- Yi SQ, Ohta T, Tsuchida A, et al. Surgical anatomy of innervation of the gallbladder in humans and Suncus murinus with special reference to morphological understanding of gallstone formation after gastrectomy. World Journal of Gastroenterology 13 (2007): 2066-2071.
- Dai Y, Ren K, Kurosawa K, et al. The distribution of nerves supplying the testis, epididymis and accessory sex glands of Suncus murinus. Anatomical Science International 94 (2019): 128-135.
- Chen S, Zhou Y, Chen Y, et al. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34 (2018): 884-890.
- Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27 (2011): 2957-2963.
- Li W, Fu L, Niu B, et al. Ultrafast clustering algorithms for metagenomic sequence analysis. Briefings in Bioinformatics 13 (2012): 656-668.
- Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nature Methods 7 (2010): 335-336.
- Mukhopadhya I, Hansen R, El-Omar EM, et al. IBD-what role do Proteobacteria play? Nature Reviews Gastroenterology & Hepatology 9 (2012): 219-230.
- Fang W, Fang Z, Zhou P, et al. Evidence for Lignin Oxidation by the Giant Panda Fecal Microbiome. Plos One 7 (2012): e50312.
- Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nature Reviews Microbiology 14 (2015): 20-32.
- Ley RE, Lozupone CA, Hamady M, et al. Worlds within worlds: evolution of the vertebrate gut microbiota. Nature Reviews Microbiology 6 (2008): 776-788.
- Pessione E. Lactic acid bacteria contribution to gut microbiota complexity: lights and shadows. Front. Cell. Infection Microbiology 2 (2012): 86.
- Hanchi H, Mottawea W, Sebei K, et al. The genus enterococcus: Between probiotic potential and safety concerns-an update. Frontiers in Microbiology 3 (2018): 1791.
- Zapasnik A, Sokolowska B, Bryla M. Role of lactic acid bacteria in food preservation and safety. Foods 11 (2022): 1283.
- Lebreton F, Willems RJL, Gilmore MS. Enterococcus diversity, origins in nature, and gut colonization. In: Gilmore MS, Clewell DB, Ike Y, et al. editors. Enterococci: From commensals to leading causes of drug resistant infection [Internet]. Boston: Massachusetts eye and ear infirmary (2014).
- Sheh A, Fox JG. The role of the gastrointestinal microbiome in Helicobacter pylori pathogenesis. Gut Microbes 4 (2013): 505-531.


 Impact Factor: * 3.8
Impact Factor: * 3.8 Acceptance Rate: 77.96%
Acceptance Rate: 77.96%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 