Optimization of Protease and Amylase Activities Derived from Soy Sauce Koji
Nguyen Xuan Hoang1, Ching-Hua Ting2, Cheng-Kuang Hsu1*
1Department of Food Science, National Chiayi University, Chiayi City, Taiwan (ROC)
2Department of Mechanical and Energy Engineering, National Chiayi University, Chiayi City, Taiwan (ROC)
*Corresponding Author: Cheng-Kuang Hsu, Department of Food Science, National Chiayi University, No.300 Syuefu Rd., Chiayi City 60004, Taiwan (ROC)
Received: 26 May 2020; Accepted: 09 June 2020; Published: 04 August 2020
Article Information
Citation:
Nguyen Xuan Hoang, Ching-Hua Ting, Cheng-Kuang Hsu. Optimization of Protease and Amylase Activities Derived from Soy Sauce Koji. Journal of Food Science and Nutrition Research 3 (2020): 153-170.
DOI: 10.26502/jfsnr.2642-11000046
View / Download Pdf Share at FacebookAbstract
The present study was aimed to determine the optimal fermentation conditions for the koji production and to evaluate the influences of temperature, pH, and brine concentration on the enzyme activity and stability of protease and amylase. The experiments were conducted by inoculating the different ratios of soybean to wheat with the spores of Aspergillus oryzae. The ratios of soybean to wheat were 2:1 and 1:1 in Chinese-type (BA1) and Japanese-type (BA2), respectively. The optimum time of koji-making for both BA1 and BA2 was found at 60 h fermentation. No remarkable difference of enzymatic characteristic was observed between BA1 and BA2, with the exception that BA1 exhibited the higher activities of both protease and amylase than those in BA2. The protease released from A. oryzae was most active and stable in the pH range 7.0-7.7 (neutral protease). The optimum temperature for protease activity was 52-55°C, and the enzyme was stable at around 40°C. Oppositely, the acidic region, pH 5, was preferred to amylase activity and stability. The amylase, which was most active at 60°C and stable at around 48°C, was more tolerant to heat and salt as compared to the protease.
Keywords
<p>Soybean sauce, Koji fermentation, Protease, Amylase</p>
Article Details
1. Introduction
Soy sauce, used as a traditional Oriental all-purpose condiment or seasoning with a salty taste and sharp flavor, is a fermented soybean food made by main raw materials including soybean, wheat and salt [1]. Generally, soy sauce can be categorized as Chinese or Japanese-type. The main difference between these two types is that Japanese-type uses soybean and wheat at a ratio of 1:1, whereas less wheat content used in Chinese-type [1-4]. The first stage of soy sauce process is koji-making, which is fermented under ~30°C with Aspergillus oryzae or A. sojae. The resultant culture mold (koji) is then mixed in an approximate 16-22% brine solution, and the obtained mash (moromi) is traditionally fermented for 4-12 months [3-5]. Finally, the refining stage includes pressing, sterilization, and packaging.
Owing to the lengthy fermentation period, unique characteristic flavor of soy sauce is artfully developed. Although the enzymatic hydrolysis is well known as dictating the quality of final product of soy sauce [5, 6, 7, 8], the characteristics of proteases and amylases, prepared using soybean and wheat fermented by A. oryzae in brine, have not been thoroughly explored. Specially, the protease and amylase characteristic derived from koji were evaluated under the changes of temperature and brine solution content [7]. As a result, the proteolytic and amylolytic activities of soy sauce koji were adversely affected by brine solution. Furthermore, the amylase was more stable to heat (45-55°C) as compared to the protease. These results as well as the suggestion of 45°C would be useful for the preparation of protein hydrolysates and biologically active peptides. Nevertheless, apart from temperature and brine solution, pH, one of the most important factors, has not yet been studied.
In the result of our previous publication, we succeed in developing the reduced-salt soy sauce product, in which the moromi mash was fermented under the low-salt environment of 10%, at 40oC for only 5 days [9]. The resultant soy sauce or enzymatic hydrolysis mash consisted of the high nutritional values of total nitrogen (> 1.4%) and amino nitrogen (> 0.56%) which met the requirement of first-class soy sauce of Taiwan [10]. As such, further study is much needed to understand the progress of enzymatic hydrolysis during the fermentation period as well as the characteristics of protease and amylase.
The objectives of the present study were to optimize the process parameters of koji production and to investigate the characteristics of enzymatic hydrolysis of koji protease and amylase, carried out from soybean and wheat fermented by A. oryzae using response surface methodology (RSM). In addition, koji prepared using the different ratios between soybean and wheat, 2:1-Chinese-type and 1:1-Japanese-type, were also studied. The results of this study would be useful for advancing the characteristics of protease and amylase and the biochemical progress during the koji making and the moromi fermentation, as well as receiving more attention with respect to the guidelines for developing preferable fermented products from koji soybean resources.
2. Materials and Methods
2.1 Raw materials and chemicals
Soybean was purchased in Taiwan and wheat was imported from Canada. The koji mold (Aspergillus oryzae) was supplied from Chuan Feng Microbe Co., Taichung City, Taiwan. All of the chemicals used in the analysis were of analytical grade.
2.2 Koji fermentation
In the koji-making stage, raw soybean was washed and then soaked in water for 24 h at 4°C and subsequently steamed at 121°C for 25 min. While wheat was roasted under 140oC for 45 min. Wheat was then cooled and cracked into smaller particles which contained 4 to 6 pieces. Steamed soybeans and roasted wheat were mixed at the different ratios, namely BA1 (Chinese-type): 2:1 (w/w) and BA2 (Japanese-type): 1:1 (w/w). The mixtures were then inoculated with 0.1% (w/w) of A. oryzae (approximately 108 spores g-1), and subsequently dispersed onto the perforated stainless steel trays (68 × 44 × 3 cm). Each tray was loaded with the mixture to a ~3 cm thickness and incubated at 30 ± 3°C. During the fermentation time, the koji was hand mixed for 5 min once every 12 h. The preparation of koji was carried out for 96 h to determine the optimal time for protease and amylase activities.
2.3 Physicochemical and microbial properties of koji during the koji making and determination of the optimal time for protease and amylase activities
At various time intervals of 12 h during the koji fermentation period, about 300 g of koji was collected after being well mixed for 5 min, then placed in sterile sampling bags, and stored at -20°C for further analysis. For each analysis, three replicates tests were performed.
Determination of microbial changes: The viable cell numbers of total bacteria and mold in samples were enumerated. Total of 5 g of each type of koji sample was added into 45 ml of sterile physiological salt solution and vortexed for 1 min. The 10-fold-concentrated cells were then serially diluted (100 to 10-6) in the same diluent, and 100 µl of properly diluted samples were spread onto plate count agar and potato dextrose agar (BD Difco, Franklin Lakes, NJ, USA) for the enumerations of total bacteria and mold, respectively. The colonies formed (25-250), counted after 2 days of incubation at 30oC, were calculated as log CFU per g koji solid [5, 11].
Enzyme extraction: Exactly 20 g of koji was mixed with 200 ml of deionized water and blended for 45s and then extracted at 30oC for 1 h with agitation at 150 rpm using sharking incubator. The homogenate was then centrifugated at 10,000 g, 4oC for 10 min and the supernatant was collected as crude enzyme extract [5, 11].
Protease activity assay: Casein (2%, w/v) dissolved in 0.05 M buffer at desirable pH (sodium phosphate buffer for pH 7.0). A mixture of 5 ml of casein solution and 1 ml of diluted enzyme extract was incubated at 40°C for 10 min. The enzyme stability was also determined by incubating at 40°C for 1 h. The reaction was ended by using 5 ml of 0.11 M trichloroacetic acid (TCA). Then, the filtrated supernatant (1 ml) was added to 5 ml of 0.5 M sodium carbonate and 1 ml of Folin–Ciocalteu’s phenol reagent (1:3 diluted in water) and the mixture was kept for 30 min. The absorbance was read against a blank at 660 nm. One unit of protease activity (U) was defined as the amount of enzyme that yields the color equivalent to 1 µM of tyrosine per gram of koji solid per minute under assay conditions [6, 12, 13].
Amylase activity assay: The a-amylase activity of koji was determined based on the 3,5-dinitrosalicylic acid (DNS) method [14] with little modification. Soluble starch (3%, w/v) was dissolved in 0.05 M buffer at proper pH (sodium citrate buffer for pH 5.0). A mixture of 1 ml of 3% soluble starch solution and 1 ml of diluted enzyme extract was kept at 40°C for 10 min. The enzyme stability was also determined by incubating at 40°C for 1 h. Thereafter, 1 ml of DNS reagent, contained 1% DNS, 2% NaOH and 30% sodium potassium tartrate in distilled water, was added. The mixture was then boiled at 100°C for 15 min. The heated sample was immediately cooled to room temperature, and mixed with 9 ml of distilled water. The absorbance was measured using a spectrophotometer at 540 nm. A standard cure was made using a 0.4% w/v glucose solution. One unit of amylase activity (U) was defined as the amount of enzyme needed to release 1 µM of glucose per gram of koji solid per minute under assay conditions.
Determinations of pH and moisture content: Five gram of koji was blended with 50 ml of deionized water for 45s and the pH were measured directly with a PB-10 pH meter (Sartorius, Göttingen, Germany) [5]. The moisture content of koji was measured according to AOAC (2000) [15].
2.4 Effects of pH, temperature and salt content on the activities and stabilities of koji protease and amylase using RSM
The influences of independent variables on response factors was performed using a face-centered central composite design. The independent variables applied in experimental design were pH (5, 7 and 9), temperature (40, 50 and 60oC) and salt content (0, 8 and 16% w/v), corresponding to coded levels -1, 0 and 1, respectively. Response factors were protease activity (U/g koji solid) and amylase activity (U/g koji solid), as well as their enzyme stabilities (Table 1). The effect of pH on the enzyme activity and stability was evaluated using different buffers. In particular, pH 5.0, 7.0, and 9.0 were adjusted by using sodium citrate, sodium phosphate, and sodium carbonate buffer, respectively. Whereas, when salt content (brine solution) was considered as a variable, different salt contents were added in the substrate solutions (casein solution for protease and soluble starch for amylase activity). The different temperatures of enzyme reactions were controlled by a sharking incubator (± 0.1oC).
The following second order quadratic equation was used to describe the correlation between the responses and the independent variables (using coded levels).
Quality response functions:
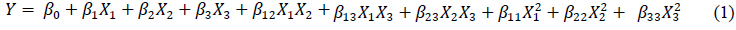
where Y was response variable. Namely, P1-10, P1-60, A1-10 and A1-60 were the protease activities with 10 and 60 min reactions and the amylase activities with 10 and 60 min reactions in BA1, respectively; P2-10, P2-60, A2-10 and A2-60 were the protease activities with 10 and 60 min reactions and the amylase activities with 10 and 60 min reactions in BA2, respectively. X1, X2 and X3 represented the independent variables for pH, temperature (°C) and salt content (% w/v), respectively. β0 was the intercept; β1-3 and β11-33 were the linear and quadratic regression coefficients, respectively; β12, β13 and β23 were the interactive regression coefficients. Analysis of variance (ANOVA) was used to evaluate model adequacy and statistical significance. The adequacy of model was evaluated by the determination coefficient (R2) and model p-value. 3-D response surface plots were performed by using Design-Expert software 10.0.1 (Stat-Ease, Minneapolis, MN, USA) for understanding the interactive influences of the independent variables on the responses. A commercial optimization software, AMPL (The Scientific Press, San Francisco, CA, USA), was used to search the optimum pH (X1), temperature (X2), and salt content (X3) to maximize the activities and stabilities of protease and amylase, particularly P1-10, P1-60, A1-10, A1-60, P2-10, P2-60, A2-10 and A2-60.
2.5 Statistical analysis
All tests were conducted in triplicate. The data were analyzed by analysis of variance (ANOVA), and significant differences in mean values among data were determined at p< 0.05 by Duncan’s multiple-ranges test using SPSS 20 (SPSS Inc., Chicago, IL).
3. Results and Discussion
3.1 Physicochemical and microbial properties of koji during the koji making and the determination of the optimal fermentation time for protease and amylase activities
Figure 1 presents the changes in the moisture content and microbial count in koji during koji-making. At the beginning of koji making-0 h, the moisture content of BA1 was 49.12%, which was higher as compared to BA2, 46.34%. Then, the moisture contents decreased sharply over time throughout the koji fermentation in both samples, in which the higher figures were also observed in BA1. The results were in accordance with the studies of Chancharoonpong, Hsieh, & Sheu [16, 17]. The higher moisture content in BA1 could be explained based on the higher amount of soybean used in koji making.
Regarding the changes of microorganism counts of different koji, the similar trends and no marked differences in total viable and mold counts were noted between two types of koji, BA1 and BA2. The period from 0 to 24 h could be considered as lag and log phases, while the remaining time were stationary phases. It was observed that the numbers of total viable counts were similar to those of mold in both samples, indicating that only few of bacteria contaminated in koji making as well as the most of survival cells were mold, particularly A. oryzae [16].
The changes of protease and amylase activities during 96 h koji fermentation are shown in Figure 2a and 2b. The protease and amylase activities were about 0 U/g koji solid (U) at the initial fermentation of 12 h, then the protease activities dramatically increased and reached a peak of ~4200 U at 60 h and significantly decreased thereafter in both BA1 and BA2. While the amylase activities remarkably rose until the period between 48 and 72 h, then tend to plateau to the subsequent fermentation. Furthermore, with the exception of protease at 60 h, the higher protease and amylase activities were observed in BA1 during the fermentation time, which might be due to the ratio of substrates used in BA1 was more favored to A. oryzae producing protease and amylase. Although the figures for the growth curves of mold showed no marked difference between BA1 and BA2 (Figure 1b, 1c).
The initial pH values of BA1 and BA2 were 6.53 and 6.54, respectively, decreased until 24 h, and then moderately went up thereafter (Figure 2c). Additionally, BA1 showed the higher pH values than those of BA2 over time throughout the fermentation with the exception of beginning fermentation of 12 h. The enhancement of pH after 24 h of cultivation period were because of the productions of various enzymes from A. oryzae [5, 11, 16], which could be seen from the results of enzyme activities after 24 h (Figure 2a, 2b). Consequently, the higher protease and amylase activities resulted in increasing pH values in BA1 as compared to BA2.
Based on these results, the optimal balance of protease and amylase activities was obtained at 60 h koji fermentation for both BA1 and BA2. Therefore, we selected the koji fermented at 60 h for the subsequent experiments.
3.2 Effects of pH, temperature and salt content on the activities and stabilities of koji protease using RSM
Table 1 shows the experimental data using the three-factor face-centered central composite design. The experiment results were fitted to a second-order polynomial model as describe in Eqn. (1). The resulting model parameters are presented in Table 2. According to particularly low p-values associated with F test for all models indicated that the models were highly significant. For all responses, the adequacy of fit of the models were strongly confirmed by the high R2 values (0.85-0.98).
Table 2 displays that only linear term of salt content exhibited significant influence on P1-10 and P2-10, which was negative effect in both cases, meaning that protease activities in BA1 and BA2 decreased with increased salt content. Furthermore, only the quadric term of pH exhibited negative effect on P1-10 and P2-10, which means that protease activities increased with pH’s increase, reached maximum at pH around 7.0-7.7 and then started to decrease as continuous increase of pH (Figure 3a, 3c). Whereas all the interaction terms were not statistically significant for protease activities measured with 10 min reaction. The similar properties were observed between P1-10 and P2-10, however, P1-10 had the higher values than P2-10. These results were consistent with what were observed during the koji fermentation (Figure 2a).
Results of earlier studies found that the neutral and alkaline proteases mainly contributed to the characteristic of koji protease, prepared using soybean and wheat fermented by A. oryzae [2, 6, 8, 13]. Thus, these finding were therefore confirmed under our investigation. Opposite results were, nevertheless, observed in the report of Janser, Castro, & Sato [18], where they stated that the protease from A. oryzae LBA 01 was more active in the pH range 5.0-5.5. This might be because the differences of koji fermentation employed, of particular for instance, the use of different A. oryzae strain and different koji substrate.
The result of P1-60 was similar to that of P2-60, in which P1-60 also showed the higher activity. All variables exhibited significant influences on the protease activities, which were linear terms of temperature and salt content and quadratic term of pH (Figure 3b, 3d). The protease activities were also more active in the neutral pH values. This indicated that protease activity was more stable at low salt content (<10%) and temperature of 40°C. Hence, results of the present study were in good agreement with our previous study, stating that the optimal conditions of enzymatic treatment for total and amino nitrogen contents were the salt content of 10% and the temperature of 40°C [9].
Accordingly, P1-10 and P2-10 showed no significant effects of temperature applied in the range of 40-60°C on the protease activities. This might be because the effect of temperature on the protease activity was not high enough within very short reaction time of 10 min. This could be seen from the results of P1-60 and P2-60 as the reaction time was longer (60 min), the temperature was significant variable, however, its contributions to protease activities were still much lower than those of pH and salt content, which were partly reflected by the regression coefficients of temperature (linear) were much lower than those of pH (quadratic) and salt content (linear) (Table 2). Hence, in the treatment of elevated temperature (40-60°C), the temperature of 40°C should be considered as an effective application for high protease activity as well as economic efficiency. Besides, without salt was preferable or the range of salt content 5-10% for the consideration of microbial contamination of enzymatic hydrolysis.
Salt content and pH played the crucial roles contributing to the protease activities. Especially, salt content could significantly explain 66% ((-0.81)2), 58% ((-0.76)2), 61% ((-0.78)2) and 44% ((-0.66)2) total variances of P1-10, P1-60, P2-10 and P2-60, respectively (Figure 4). This result confirmed that salt content was one of the most vital factors in defining the aging period and qualities of soy sauce [2, 19, 20, 21]. In addition, it could be seen from run 5 and 6 (Table 1) that the proteases had abilities to adapt to the salt stress since the activities significantly increased in the period from 10 (P1-10 and P2-10) to 60 min reactions (P1-60 and P2-60). Besides, the higher stabilities of proteases at 40°C also contributed to the increased activities in run 5 and 6 as the protease activities in run 7 and 8 declined with the temperature reaction of 60°C.
In the comparation between BA1 and BA2, apart from higher protease activities were observed in BA1, there was no marked difference of the protease characteristic. This could be explained by good correlations detected in Figure 4, namely P1-10 and P2-10 (r = 0.96), P1-60 and P2-60 (r = 0.98).
3.3 Effects of pH, temperature and salt content on the activities and stabilities of koji amylase using RSM
Figure 5 shows similar shapes between A1-10 and A2-10, and A1-60 and A2-60. In addition, the good correlations, which had significant coefficients of correlations r = 0.99 (Figure 4), were also observed between them. Therefore, the characteristic of amylase of BA1 was also similar to that of BA2, with the only difference was the higher figure in BA1.
According to results from Table 2, pH and temperature showed significant influences on the amylase activities of A1-10 and A2-10, which was negative effect for pH but positive influence for temperature. This means the increased pH and/or decreased temperature would cause the decrease of amylase activities (Figure 5a, 5c). As a result, the amylases were more active at acidic regions (pH 5.0). Surprisingly, salt content had no effects on the amylase activities, and interaction terms with another variable as well (Table 2 and Figure 5a, 5c).
Nevertheless, with 60 min reaction, only linear term of pH exhibited significant influence on A1-60 and A2-60, which was negative in both cases, meaning that amylase activities increased as decreased pH. Salt content and temperature had no influences on both A1-60 and A2-60, also their interaction terms with another variable did not show influential effect. Although temperature had no significant influences on the models of A1-60 and A2-60, the effects of temperature on amylase activities tend to change from linear terms (A1-10 and A2-10) to quadratic terms (A1-60 and A2-60) with the optimum ranges of 45-50°C (Figure 5b, 5d).
Moreover, in run 2, 13, 14, 15, 16 and 17 (Table 1), where the temperatures applied were 40-50°C and although the treatments of pH 7.0-9.0 were not favored to the amylases, the amylases activities significantly increased in the short period between 10 (A1-10 and A2-10) and 60 min reactions (A1-60 and A2-60). Thus, the amylases probably had abilities to adapt to the pH stress. Based on the mechanism of “lock-key theory” for enzymatic reaction [22], we hypothesized that at the beginning of 10 min reaction, the inhibitors of high salt content and neutral-alkaline regions of pH suddenly changed the shapes of protease’s and amylase’s active sites, respectively, and made them unusable to the substrates, resulting in the significant reductions in the reaction rate. Nevertheless, as the rose time of 60 min, the bindings of these altered shapes of enzyme’s active sties and intended substrates were more and more formed, the rate of reactions were therefore increased corresponding to the increased enzymes activities of protease and amylase.
The results showed that pH played the most important role contributing to the changes of amylase activities and the amylase was more active at acidic pH region around 5.0. This was in good agreement with the results of previous studies that fungal amylases were generally stable in pH range 2.0-6.0 [12, 23]. Furthermore, fungal amylase of A. oryzae in present study had the higher abilities of thermostable and high salt-tolerant than those of protease. In agreement, Su et al. [7] also found that the protease was more sensitive to heat as compared to the amylase.
3.4 Optimization
A nonlinear programming solver, AMPL, was used to search the optimum pH, temperature, and salt content to maximize P1-10, P1-60, A1-10, A1-60, P2-10, P2-60, A2-10 and A2-60 presented in Table 3. As a result, the optimal conditions of pH were neutral regions (7.0-7.7) for protease activities and acidic regions (5.0) for amylase activities in both of BA1 and BA2. Whereas without salt or very low salt contents (5.17% for A1-10 and 3.69% for A2-10) were generally optimized. Besides, the temperatures exceeded 52°C promoting the enzymatic hydrolysis within 10 min reaction, P1-10, A1-10, P2-10 and A2-10. However, when the thermal stability was considered, the lower temperatures were preferable, namely around 40°C for proteases (P1-60 and P2-60) and approximately 48°C for amylases (A1-60 and A2-60). Protease and amylase activities at these optimum conditions were also determined to validate the regression models, and the results showed that the experimental values agreed well with the values predicted by the models.
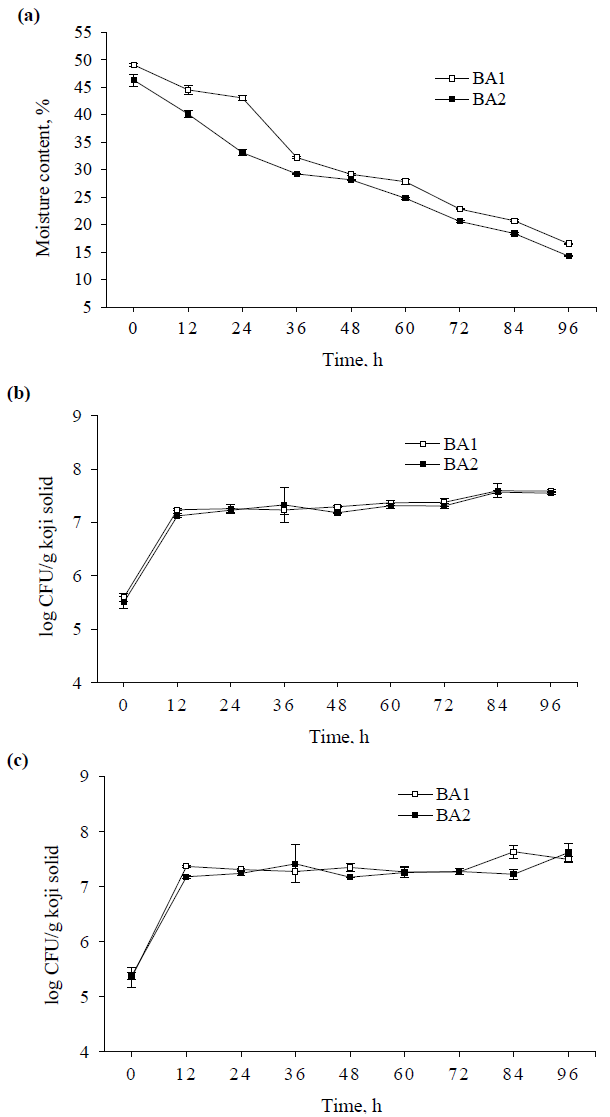
Figure 1: (a) Changes of moisture content; (b) total viable count; and (c) total mold of different koji at various periods of koji fermentation.
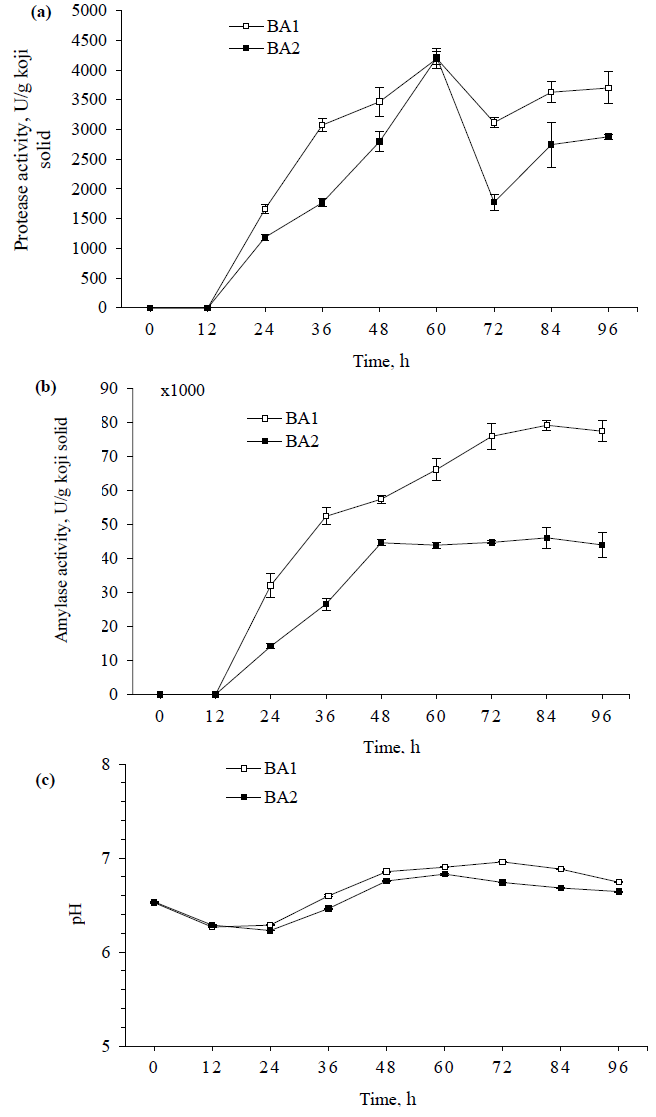
Figure 2: (a) Changes of protease activity; (b) amylase activity; and (c) pH of different koji at various periods of fermentation.
Figure 3: The response surface plots of protease activity influenced by the reaction parameters of pH, temperature and salt content using coded values; (a, b) Protease activities of BA1 with 10 (P1-10) and 60 min reactions (P1-60), respectively; (c, d) Protease activities of BA2 with 10 (P2-10) and 60 min reactions (P2-60), respectively.
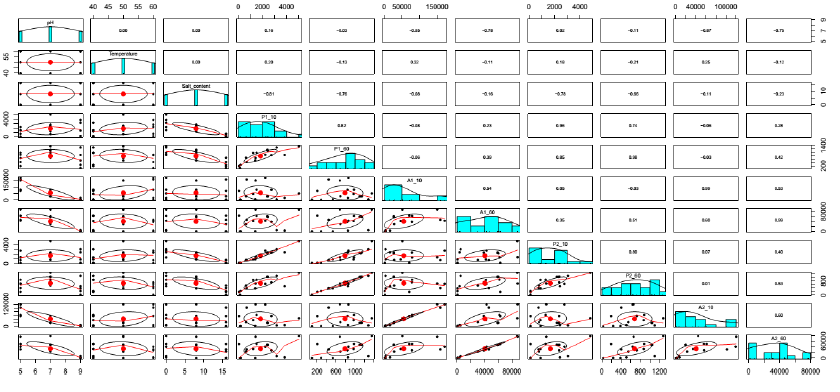
Figure 4: Correlation between independent variables and responses.
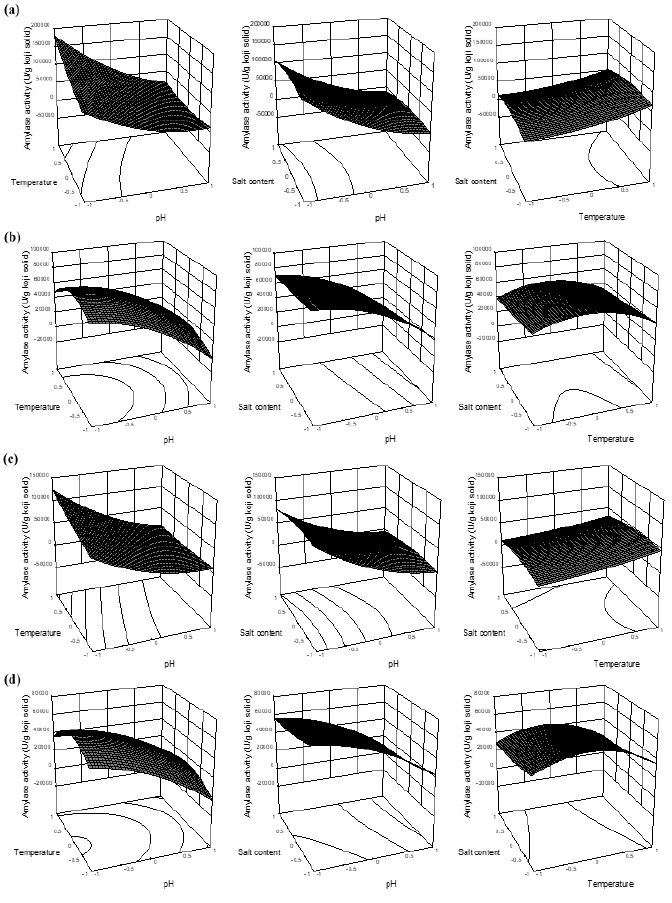
Figure 5: The response surface plots of amylase activity influenced by the reaction parameters of pH, temperature and salt content using coded values. (a, b) Amylase activities of BA1 with 10 (A1-10) and 60 min reactions (A1-60), respectively; (c, d) Amylase activities of BA2 with 10 (A2-10) and 60 min reactions (A2-60), respectively.
P1-10, P1-60, A1-10 and A1-60 were the protease activities with 10 and 60 min reactions and the amylase activities with 10 and 60 min reactions in BA1, respectively; P2-10, P2-60, A2-10 and A2-60 were the protease activities with 10 and 60 min reactions and the amylase activities with 10 and 60 min reactions in BA2, respectively.
Table 1: The treatment combinations and their responses.
*Significant at 0.05 level; aCoded values; bCoefficient of determination
Table 2: Summary of regression analysis.
*Data are expressed as the mean ± SD (n = 3).
Table 3: Optimal conditions and model validation.
4. Conclusions
The optimum of koji-making time for both BA1 and BA2 was 60 h. Besides, BA1 and BA2 had the same properties of protease and amylase. The protease produced by A. oryzae was most active and stable in the pH range 7.0-7.7, the situation of neutral protease. The optimum temperature for protease activity was 52-55°C, and the enzyme was stable at around 40°C. On the contrary, the acidic region, pH 5, was preferred to amylase activity and stability. Additionally, the amylase, which was most active at 60°C and stable at around 48°C, was more tolerant to heat and salt as compared to the protease. The appearance of salt adversely affected the protease and amylase activities, resulting in the optimum content of without salt for stabilities of enzymes. Salt content was the most crucial factor contributing to the characteristic of protease, while pH value for amylase. Besides, the protease and amylase had adapt-abilities to salt stress and pH stress, respectively. Our results suggest that the conditions of pH 7.0-7.7 and temperature of 40°C, and the conditions of pH 5.0 and temperature of 48°C should be applied in the food industry as the methods of choice to quantify soy sauce koji protease and amylase activities, respectively, derived from A. oryzae.
Acknowledgements
This research was supported by the Ministry of Science and Technology in Taiwan, R.O.C. (MOST 105-2320-B-415-005-).
References
- Zhang D, He Y, Cao Y, et al. Flavor improvement of fermented soy sauce by extrusion as soybean meal pretreatment. Journal of Food Processing and Preservation 41 (2017): 1-8.
- Huang TC, Teng DF. Soy Sauce: Manufacturing and biochemical changes. In Eds.: Hui YH, Meunier-Goddik L, Hansen AS, et al. Handbook of food and beverage fermentation technology. New York: CRC Press (2004).
- Kaneko S, Kumazawa K, Nishimura O. Comparison of key aroma compounds in five different types of Japanese soy sauces by aroma extract dilution analysis (AEDA). Journal of Agricultural and Food Chemistry 60 (2012): 3831-3836.
- Van Der sluis C, Tramper J, Wijffels RH. Enhancing and accelerating flavour formation by salt-tolerant yeasts in Japanese soy-sauce processes. Trends in Food Science and Technology 12 (2001): 322-327.
- Liu J, Li D, Hu Y, et al. Effect of a halophilic aromatic yeast together with Aspergillus oryzae in koji making on the volatile compounds and quality of soy sauce moromi. International Journal of Food Science and Technology 50 (2015): 1352-1358.
- Chen ZY, Feng YZ, Cui C, et al. Effects of koji-making with mixed strains on physicochemical and sensory properties of Chinese-type soy sauce. Journal of the Science of Food and Agriculture 95 (2015): 2145-2154.
- Su NW, Wang ML, Kwok KF, et al. Effects of temperature and sodium chloride concentration on the activities of proteases and amylases in soy sauce koji. Journal of Agricultural and Food Chemistry 53 (2005): 1521-1525.
- Yong FM, Wood BJB. Microbiology and biochemistry of soy sauce fermentation. Advances in Applied Microbiology 17 (1974): 157-194.
- Hoang NX, Ferng S, Ting CH, et al. Optimizing the initial moromi fermentation conditions to improve the quality of soy sauce. LWT - Food Science and Technology 74 (2016): 242-250.
- Chinese National Standard (CNS). Soy Sauce-CNS423, N5006 (2002).
- Bechman A, Phillips RD, Chen J. Changes in selected physical property and enzyme activity of rice and barley koji during fermentation and storage. Journal of Food Science 77 (2012): 318-322.
- Negi S, Banerjee R. Optimization of amylase and protease production from Aspergillus awamori in single bioreactor through EVOP factorial design technique. Food Technology and Biotechnology 44 (2006): 257-261.
- Su G, Ren J, Yang B, et al. Comparison of hydrolysis characteristics on defatted peanut meal proteins between a protease extract from Aspergillus oryzae and commercial proteases. Food Chemistry 126 (2011): 1306-1311.
- Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Analytical Chemistry 31 (1959): 426-428.
- Official Methods of Analysis (17th ed.). Gaithersburg, Maryland: Association of Official Analytical Chemists (2000).
- Chancharoonpong C, Hsieh PC, Sheu SC. Production of enzyme and growth of Aspergillus oryzae S. on soybean koji. International Journal of Bioscience, Biochemistry and Bioinformatics 2 (2012a): 228-231.
- Chancharoonpong C, Hsieh PC, Sheu SC. Effect of different combinations of soybean and wheat bran on enzyme production from Aspergillus oryzae S. APCBEE Procedia 2 (2012b): 68-72.
- Janser R, Castro SD, Sato HH. Protease from Aspergillus oryzae: biochemical characterization and application as a potential biocatalyst for production of protein hydrolysates with antioxidant activities. Journal of Food Processing 2014 (2014): 1-11.
- Gao X, Cui C, Ren J, et al. Changes in the chemical composition of traditional Chinese-type soy sauce at different stages of manufacture and its relation to taste. International Journal of Food Science and Technology 46 (2011): 243-249.
- Song YR, Jeong DY, Baik SH. Effects of indigenous yeasts on physicochemical and microbial properties of Korean soy sauce prepared by low-salt fermentation. Food Microbiology 51 (2015): 171-178.
- Hoang NX, Ferng S, Ting CH, et al. Effect of initial 5 days fermentation under low salt condition on the quality of soy sauce. LWT - Food Science and Technology 92 (2018): 234-241.
- Whitehurst RJ, Van Oort M. Enzymes in food technology (2nd Edn.) Iowa: Wiley-Blackwell (2010).
- Ominyi MC. Optimization of α-amylase and glucoamylase production from three fungal strains isolated from Abakaliki, Ebonyi State. European Journal of Experimental Biology 3 (2013): 26-34.


 Impact Factor: * 3.8
Impact Factor: * 3.8 Acceptance Rate: 77.96%
Acceptance Rate: 77.96%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 