Microbial Community and Physicochemical Characterization of Kombuchas Produced and Marketed in Brazil
Taís Suhre1#, Michele Bertoni Mann2#, Caroline Isabel Kothe3, Artur Luiz Guedes Rocha4, Paulo Gustavo Celso4, Ana Paula Muterle Varela2,5, Ana Paula Guedes Frazzon2, Jeverson Frazzon1*
1Federal University of Rio Grande do Sul (UFRGS), Institute of Food Science and Technology (ICTA), Porto Alegre, RS, Brazil
2UFRGS, Institute of Healthy Basic Science (ICBS), Porto Alegre, RS, Brazil
3Université Paris-Saclay, INRAE, AgroParisTech, Micalis Institute, Jouy-en-Josas, France
4Ministry of Agriculture, Livestock, and Supply (MAPA), Laboratory of Food Identity and Quality (LFDA-RS), Porto Alegre, RS, Brazil
5Federal University of Health Sciences of Porto Alegre (UFCSPA), Porto Alegre, RS, Brazil
*Corresponding Author: Jeverson Frazzon, Federal University of Rio Grande do Sul (UFRGS), Institute of Food Science and Technology (ICTA), Porto Alegre, RS, Brazil.
Received: 16 November 2021; Accepted: 23 November 2021; Published: 03 December 2021
Article Information
Citation:
Suhre T, Mann MB, Kothe CI, Rocha ALG, Celso PG, Varela APM, Frazzon APG, Frazzon J. Microbial community and physicochemical characterization of kombuchas produced and marketed in Brazil. Journal of Food Science and Nutrition Research 4 (2021): 302-316.
DOI: 10.26502/jfsnr.2642-11000082
View / Download Pdf Share at FacebookAbstract
Abstract Kombucha has recently become popular in the Brazilian beverage market as a healthy alternative to soft drinks. However, little is known about the microbial composition and physicochemical characteristics of products available on the market. To investigate the microbial profile of kombuchas, high-throughput sequencing of the 16S rRNA and ITS genes, in samples belonging to six brands was utilized. In addition, the drinks were characterized based on the physicochemical parameters of pH, total acidity and alcohol content. Through the metagenetic analysis, the most abundant prokaryotic species identified were Liquorilactobacillus nagelii, Oenococcus oeni, Komagataeibacter rhaeticus, Liquorilactobacillus ghanensis, Gluconobacter oxydans, Komagataeibacter saccharivorans, Acetobacter peroxydans and Pantoea stewartii, while the mainly eukaryotic species were Dekkera bruxellensis, Dekkera anomala, Saccharomyces cerevisiae and Lanchancea fermentati. Interestingly, we identified six different oligotypes of D. bruxellensis, showing a wide diversity of strains belonging to this species. The results obtained for the physicochemical analyses, within the shelf life of the products, presented a range between 2.88 ± 0.06 and 3.43 ± 0.04 of pH, values between 1.80 ± 0.59 and 4.86 ± 0.72 for the total titratable acidity and 1.03 ± 0.24 to 2.54 ± 0.39 referring to alcohol content, demonstrating significant differences between brand. In addition, all samples had alcohol content above 0.5%, resulting in the classification of alcoholic beverages, which need proper labelling. The data generated in this work helped to understand the composition of the kombuchas available in the Brazilian market, as well as in the development of the identity and quality standard of the drink.
Keywords
<p>Beverage, Black Tea, Fermentation, Green Tea, Kombuchas, Legislation</p>
Article Details
1. Introduction
During the past few decades, kombucha has transitioned from a homemade fermented drink to a commercially and industrially produced drink. According to Kombucha Brewers International [1], this is the product with the highest growth estimate in the beverage market [1]. Additionally, while in the United States, a strong growth rate of 17.5% is expected for this market between 2019 and 2024 [2], in Brazil, this sector is quite new. The first factory was established in 2016 and the Brazilian Association of Kombucha, founded in 2018, already includes approximately 25 associated producers [3]. The diffusion of kombucha is mainly attributed to its supposed functional properties, particularly related to the production of organic acids, as well as the presence of probiotic microorganisms and polyphenols [4]. However, although the beverage is considered a miracle cure for a variety of diseases, according to popular wisdom [5], there is little clinical evidence about its prophylactic and beneficial effects on human health [6,7]. In addition, information about the physicochemical and microbial composition of commercial samples is unknown to consumers and are important especially for risk groups such as pregnant women, drivers, children and immunocompromised patients [8,9]. Slightly sweet and acidic, the drink is produced from the fermentation of several substrates [10]. Black or green teas (Camellia sinensis) are the most used [7]. After preparing the tea in boiled water and adding sucrose, the initial fermentation takes place at room temperature and aerobic conditions [4]. Fermentation is carried out from an initial culture containing bacteria and yeasts, known as SCOBY (Symbiotic Culture of Bacteria and Yeasts). The process also involves adding 10 to 20% of a previously fermented kombucha to acidify the medium and prevent the growth of pathogenic microorganisms [11]. After this fermentation, which takes approximately 7 to 10 days, the resulting product can be refrigerated and consumed [12]. However, the kombucha can still undergo a second fermentation. At this stage, a new source of sugar is added. Usually, the sources used are juice, fruit or sucrose. Thus, natural carbonation occurs in the bottle, giving the drink a clear and sparkling characteristic. Forced carbonation of the kombucha can also occur through the incorporation of carbon dioxide. During kombucha fermentation, the invertase enzymes produced by some yeasts cleave sucrose into glucose and fructose. In turn, yeasts convert glucose into ethanol and carbon dioxide [13]. Meanwhile, bacteria oxidize some of the ethanol to produce acetic acid, lowering the pH of the medium. Moreover, several other metabolites, such as gluconic, lactic, tartaric and citric acid, are produced, which may contribute to the drink's characteristic flavor. However, the composition and concentration of the metabolites depends on the source of SCOBY, the concentration of sugar and tea, the fermentation time and the temperature used. Thus, any change in fermentation conditions can affect the organoleptic and physicochemical characteristics of the final product [12]. It was reported that the microbial composition of SCOBY can vary between fermentations, according to its origin, climate, geographic location and medium used for the fermentation process [8,14]. However, many studies on the kombucha microbiome are carried out through microbial culture-dependent analysis, which can produce ambiguous results due to the variability of the species. In addition, samples can contain non-cultivable microorganisms, making the description of the community unreliable [11]. Thus, DNA sequencing methods, such as those used in this work, are efficient tools for a better understanding of the microbial communities present in the products found on the shelves.
Therefore, due to the rapid expansion of kombucha in the Brazilian market and because it is considered a "healthy" substitute for soft drinks, understanding the composition of this product becomes particularly important. In this context, the aim of the present study was to evaluate the microbiological and physicochemical profile of kombuchas produced and commercialized in Brazil. Samples from six brands were analyzed using High-Throughput Sequencing (HTS) of the ITS and 16S rRNA genes. In addition, the physicochemical parameters, such as total titratable acidity, pH and ethanol content of the drinks, were determined.
2. Materials and Methods
2.1 Experimental design and sampling
Six brands of kombucha produced and marketed in Brazil were evaluated in the present study within the shelf life of the products (Figure 1). The samples were kindly provided by the manufacturers and included three brands of unflavored kombuchas (original flavor) produced with green tea base (K1, K3 and K4), one brand of unflavored kombucha (original flavor) with black tea base (K5) and two brands of kombuchas flavored with grapes, one produced with a base of green tea (K6) and the other with a mixed base of green tea and mate tea (K2). The samples were transported to the Laboratory of Biochemistry and Molecular Biology of Microorganisms (LBMBM / UFRGS) in their original packaging and kept refrigerated at 4 °C until pre-processing prior to analysis.
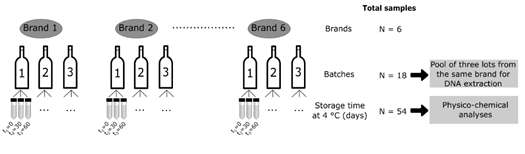
Figure 1: Schematic diagram of the sampling method and analyses.
2.2 Sample processing
For microbiome analysis, a volume of 50 mL of each batch sample was centrifuged at 4,000 rpm for 40 minutes. The supernatants were discarded and the pellets formed for each brand were grouped in pools and subjected to total DNA extraction. For physicochemical analysis, the bottles were opened carefully and the liquid was dispensed into Falcon tubes. The tubes were centrifuged for 5 minutes at 5,000 g at 4 °C. Then, the collected supernatant was frozen in liquid nitrogen and stored in an ultra-freezer (-80 °C). The processing of the samples was carried out at the LBMBM/UFRGS.
2.3 DNA extraction
For bacterial characterization, the DNA of samples were extracted using the DNeasy Blood & Tissue kit (QIAGEN, Hilden, Germany). For fungal characterization, the total DNA was extracted using the E.Z.N.A.® Stool DNA Kit (Omega Bio-tek). The extracted DNAs were quantified using a Qubit ® 3.0 fluorimeter (INVITROGEN, California, USA), and used for partial amplification of the 16S rRNA and ITS genes through Polymerase Chain Reaction (PCR). The DNA was eluted in 25 µL Milli-Q water and stored at -20 °C until the time of analysis.
2.4 Library preparation and amplicon sequencing
The 16S rRNA PCR libraries were generated by amplifying the V4 region of the 16S rRNA gene, using primers F515 and R806 [15], both modified to contain an Illumina-adapting region. The amplification was performed in a 25 µL mixture, consisting of 12.5 ng genomic DNA, 1 mM MgCl2, 0.2 mM dNTP, 0.5 μM of each primer, 2U Platinum Taq DNA Polymerase (INVITROGEN, California, USA) and 1X reaction buffer. Amplification was performed in the thermocycler (model 170-9703, MyCycler Termal Cycler, Bio-Rad) with initial denaturation for 3 minutes at 94 ºC, followed by 30 cycles of 30 seconds at 94 ºC, 30 seconds at 55 ºC and 30 seconds at 72 ºC, final extension of 5 minutes at 72 ºC. To generate the ITS PCR libraries, the ITS intergenic region was amplified using the ITS1 and ITS2 [16] primers, also modified to contain an Illumina adapting region. Amplification was performed in a 25 μL mixture consisting of 12.5 ng genomic DNA, 2.5 mM MgCl2, 0.2 mM dNTP, 0.16 μM of each primer, 1U Platinum Taq DNA Polymerase (INVITROGEN, California, USA) and 1X reaction buffer. Amplification was carried out with initial denaturation for 5 minutes at 95 ºC, followed by 35 cycles of 45 seconds at 95 ºC, 45 seconds at 56 ºC and 1 minute at 72 ºC, final extension of 10 minutes at 72 ºC. The amplicons were purified using Agencourt AMPure XP beads (BECKMAN COUTER, California, USA), following the manufacturer's instructions. The indexes were added to the DNA libraries following the manufacturer's instructions (Illumina Inc., San Diego, California, USA). The products from each sample were diluted and pooled for sequencing on the Illumina MiSeq™ system with a 500 cycle v2 kit.
2.5 Bioinformatic data analysis
The raw 16S rRNA and ITS sequences amplicons were imported into the FROGS pipeline [17] to obtain the Operational Taxonomic Units (OTUs). Then, the sequences with amplicon size from 50 to 500 bp were filtered and pooled into OTUs with SWARM [16], d=1. The chimera removal was performed with VSEARCH [18] and OTUs were filtered to keep at least 0.1% of all sequences. Taxonomic affiliations were checked using EzBiocloud database for bacteria, delimited at 98.65% identity [19], and UNITE database for yeasts. Plot composition and clustering was performed using the phyloseq and ggplot2 packages in R Studio v. 3.6.1 [20]. Sequencing data were deposited in the Sequence Read Archive of the National Centre for Biotechnology Information (NCBI), access number PRNJA611694.
2.6 Physicochemical analyses
The physicochemical analyses were performed in three batches of each brand at the Food Identity and Quality Laboratory (LFDA-RS / MAPA), where the samples were thawed and decarbonated in an ultrasonic bath for 60 minutes. The parameters of pH, total titratable acidity and ethanol content were determined. The determination of the pH of the samples was carried out by reading the digital potentiometer. The total titratable acidity of the kombucha samples was measured according to the methodology described by MAPA [21]. A 10 mL aliquot of kombucha was titrated with 0.1 N NaOH and the end point were determined by the inflection point of phenolphthalein (pH 8.2). Total titratable acidity was expressed in g of acetic acid per 100 mL of sample. The alcohol content determination was performed in an electronic densimeter coupled to a near infrared spectrophotometer (NIR) Alcolyzer ME Anton Paar DMA 4500M by the Alcolyzer Beer method for drinks with low alcohol content, with a tolerance of up to 0.3%. The results of the alcohol content were expressed as a percentage of ethanol.
2.7 Physicochemical statistical analysis
The statistics of physicochemical parameters were assessed by analysis of variance (One Way ANOVA) at p <0.05, followed by Tukey’s test or Kruskall-Wallis, with Dunn's a posteriori test (non-parametric data). The statistical program used was GraphPad InStat. For this statistical analysis, outliers were identified through online software GraphPad QuickCalcs through Grubb's test (ESD method - extreme studentized deviate) to determine if one of the listed values is a significant amount of the rest of the data.
3. Results and Discussion
3.1 Taxonomic characterization of Kombuchas by HTS
In this work, the sequences targeting the 16S rRNA gene were clustered in 22 bacterial OTUs, while the sequences targeting the ITS gene were clustered in 13 yeast/fungal OTUs, both with abundance >0.1% in the kombucha samples (Table S1). From the standpoint of microbial composition, it has been reported that kombucha provides a wide source of bacteria and yeast. Fermentation is carried out by a symbiotic association of microorganisms known as SCOBY, consisting of cellulose, acetic acid bacteria, lactic acid bacteria and yeast [8,22,23]. However, compared to bacteria, the diversity of yeasts was considerably lower. As described in the literature, certain bacteria present in kombucha, in addition to lowering the pH, have antifungal activity, which is a hypothesis to explain the result found [16].
3.2 Bacterial community
The analysis of the bacterial community indicated a predominance of the phyla Firmicutes and Proteobacteria in the kombucha samples (Figure 2A). In general, the five brands that had green tea in their composition (K1, K2, K3, K4 and K6) showed predominance of the phylum Firmicutes (between 69-99% of relative abundance), mainly represented by the Lactobacillaceae family. However, brand K5, the only one produced with black tea, was dominated with 99% relative abundance by the Acetobacteraceae family (Figure 2B), belonging to the phylum Proteobacteria. Among the genera identified in the green tea-based kombucha, the predominant ones were Liquorilactobacillus (K2, K4 and K6) and Oenococcus (K1 and K3), while sample K5 (produced with black tea) was dominated by Komagataeibacter and Gluconobacter (Figure 3C).
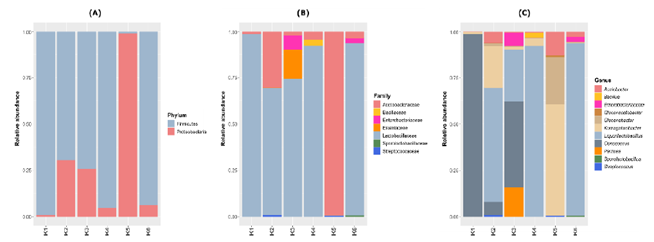
Figure 2: Bacterial plot depicting the relative abundance of the phyla, family and genera found in six Brazilian kombucha brands.
The clustering analysis separated the kombuchas in three groups with different species composition (Figure 3). The first one is composed by K1 and K3, which were dominated by Oenococcus oeni with 99% and 44% of relative abundance, respectively. We observed that the K3 was also composed by Liquorilactobacillus ghanensis (25%), Pantoea stewartii (16%) and by species belonging to Enterobacteriaceae family (7.5%). The second group is composed by K5; the only sample predominated by Proteobacteria. The mainly species presents in this kombucha were Komagataeibacter rhaeticus (44.7%), Gluconobacter oxydans (22.9%), Komagataeibacter saccharivorans (11.9%) and Acetobacter peroxydans (11.7%). Concerning the last clade, we observed three samples with similar bacterial composition: K2, K4 and K6, where the predominant species was Liquorilactobacillus nagelii (varying between 40-92%). The K2 sample was also composed mainly by Liquorilactobacillus ghanensis (21.5%), Komagataeibacter rhaeticus (11%), Komagataeibacter saccharivorans (10%), Oenococcus oeni (6%) and Acetobacter peroxydans (4.6%). Finally, the K6 was majority dominated by two species of Liquorilactobacillus. The most dominant was Liquorilactobacillus nagelii (48%) and the other belonging to an unknown species of this genus (Table S1; Figure S1).
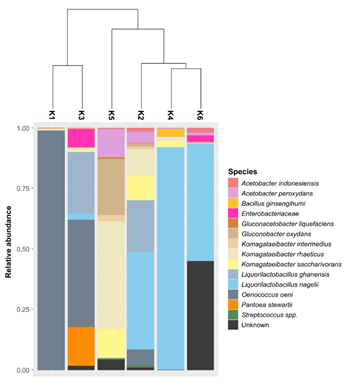
Figure 3: Clustering among bacterial OTUs identified in kombuchas and plot depicting the relative abundance of the species.
Oenococcus oeni seems to play an important, but not indispensable, role in the fermentation of kombucha. The species is not present in all samples, but has a high relative abundance in the samples in which it is present. It is often found in ciders and wines and is the main lactic acid bacteria responsible for malolactic fermentation, contributing positively to the organoleptic characteristics of wines [25,26]. Other bacteria found in both wines and kombucha is L. nagelii [27]. This species has been described in other spontaneously fermented foods and beverages such as cocoa beans, water kefir and cassava [28]. In addition, L. ghanensis was found in some kombucha brands in this study with significant relative abundance. This microorganism was also previously isolated from fermented cocoa [29]. In this sense, the presence of bacteria of the Liquorilactobacillus genus is interesting due to its potential to confer probiotic properties [30]. The presence of bacteria from the Enterobacteriaceae family was also described in other works that investigated the microbial composition of kombuchas using molecular biology methodologies [8,31]. Some species of this family naturally inhabit the intestines of mammals, while others are potential causes of disease [30]. However, the low pH of kombucha, resulting from the biosynthesis of organic acids during fermentation, prevents the development of potential pathogens [32]. On the other hand, K. rhaeticus and K. saccharivorans was detected in all kombucha samples analyzed here and as found in other studies, these bacteria are associated with cellulose production [33,34]. Finally, the bacteria G. oxydans and A. peroxydans found in this study are considered spoilage in wine and beer. However, in kombucha they play a fundamental role in the fermentation process [8,35]. These bacteria promote the oxidation of ethanol to acetic acid which is important to protect the kombucha from the growth of pathogenic microorganisms and to decrease the final ethanol concentration in the beverage.
3.3 Fungal community
For all kombucha samples, the fungal class detected was Saccharomycetales, belonging to the phylum Ascomycota (Table S1). Regarding the eukaryotic family, the dominant one was Pichiaceae (which ranged from 76.9 to 100% of relative abundance in all samples) (Figure 4A). The kombuchas K1 and K3 also had important proportions of the Saccharomycetaceae family, with 23 and 15.2% of relative abundance, respectively. The Picchiaceae family was represented mostly by the genus Dekkera, while the Saccharomycetaceae by the genera Saccharomyces spp. and Lachancea spp. in samples K1 and K3, respectively (Figure 4B). An interesting fact is that Saccharomyces cerevisiae was present with the highest significant relative abundance in the K1 brand, which was also the brand that showed the highest concentrations of ethanol in the physicochemical analyses.
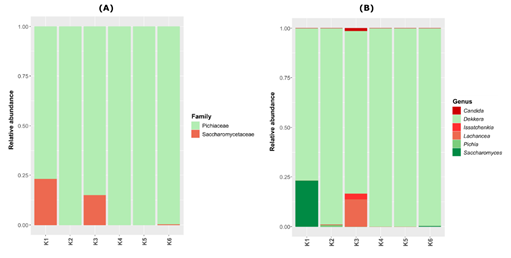
Figure 4: Fungal plot depicting the relative abundance of the genera and family found in six Brazilian kombucha brands.
Dekkera anomala and Dekkera bruxellensis were the most prevalent species, identified in all samples; however, interestingly, we identified six different oligotypes corresponding to D. bruxellensis (Figure S2; Table S1). This presence of multiple strains per species highlights the intraspecific diversity. The clustering analysis for the eukaryotic OTUs separated the kombuchas in three groups (Figure 5). The first one with only one sample (K3), that was mainly dominated by D. anomala (63.7%), followed by D. bruxellensis (Db1, 16.8%), Lachancea fermentati (13.9%) and Issatchenkia orientalis (3%). The second clade was composed by K2 and K4 samples, majority dominated by different oligotypes of D. dekkera (Db1, Db2 and Db4 in common) (Figure 5). The last group contain the K1, K5 and K6 kombuchas and is composed mainly by D. bruxellensis Db1, with amounts that varied between 27.1 to 95% (Table S1).
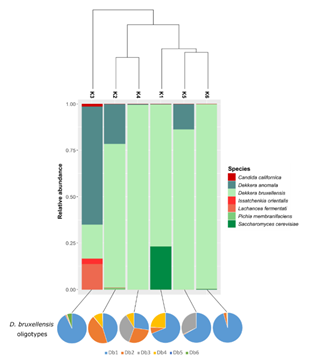
Figure 5: Clustering among fungal OTUs identified in kombuchas, barplot depicting the relative abundance of the species and pie charts showing the abundance of D. bruxellensis oligotypes.
The main species of yeasts found in this study (D. bruxellensis and D. anomala) are acid tolerant, known for their important role during the fermentation of Belgian Lambic beers. They have a positive contribution in taste through the production of acetic acid. Even so, the natural fermentation carried out from these yeasts, produces phenolic compounds, with sensory descriptors of typical aromas such as burnt plastic, horse sweat and stable. In wine, D. bruxellensis is considered one of the main deteriorating microorganisms, but it is also an aromatic profile characteristic of some products, such as the French Château de Beaucastel [36]. Predominant presence of these species in kombucha has been reported in several studies [8,37,5] and appears to play an important role in the fermentation of the drink. In the alcoholic fermentation of wine, the highest adaptation rate of D. bruxellensis was demonstrated, to survive in the must, in comparison with other wild yeasts [38]. Thus, considering the similar conditions of the two environments, this may explain the predominance of these species in most of the kombucha samples analyzed in this work. In addition, L. fermentati was detected in sample K3 and with a relative frequency <1% in sample K2. In this context, the genus Lachancea appears frequently in the fungal composition of kombucha described in other works [39,12]. It has also attracted attention in recent years, due to its unusual metabolic characteristic of being able to produce lactic acid during alcoholic fermentation. Its use is being investigated to reduce pH and increase total acidity in wine production [40,41], and to produce low alcohol beers [39]. The use of L. fermentati has also been used in the production of acidic beers, for the production of beverages without the use of bacteria or the addition of the lactic acid reagent. Although L. fermentati is considered non-pathogenic, there is a report of a case of fungemia in an immunocompromised patient, related to this species [42]. In turn, I. orientalis is a species reported in oenology, being frequently found on the surface of fruits and fruit juices. This fungus is capable of converting glucose and fructose at elevated temperatures, above those tolerated by the yeast Saccharomyces cerevisiae, thus being able to act cooperatively with this species [43].
3.4 Physicochemical analysis
There were statistically significant differences between the products of the brands in the analyzed physicochemical parameters of pH, total titratable acidity and alcohol content (Table 1). During kombucha fermentation, the pH of the tea, which initially is approximately 5, decreases due to the increase in the concentration of organic acids produced by microorganisms during the fermentation process [44,45]. In the pH analysis, performed in Brazilian kombuchas, the values found between the brands varied between 2.88 ± 0.06 and 3.43 ± 0.04, indicating that kombuchas may have different acid features that could influence in the organoleptic characteristics. Thus, these pH values found below indicate the production of organic acids, which interrupt and inhibit the proliferation of most pathogenic microorganisms [46]. In addition, this parameter is relevant due to the fact that the pH must be higher than 2.0, to avoid dental and / or gastrointestinal problems for consumers and guarantee a sensorially pleasant drink [45]. Furthermore, as evidenced in this work, kombucha fermentation resulted in the formation of ethanol, exceeding Brazil's regulatory limit of 0.5% (v / v) for non-alcoholic beverages, defined in normative statement N°41 of Ministry of Agriculture, Livestock and Food Supply [47]. However, the kombuchas analyzed had no indication of the alcohol content on their labels and were analyzed before the legislation came into effect. Likewise, in a study using gas chromatography as a methodology, in which 18 kombuchas available on the American market were analyzed, ethanol values in the range of 1.12–2.00% (v / v) were observed [9], indicating that the problem of alcohol content is not exclusive to Brazilian products. These results demonstrate the importance of adequate labelling and the development and use of quality control methodologies for these products, mainly to monitor the ethanol concentration of commercial kombuchas. As the production of the drink occurs in an aerobic environment, part of the ethanol is oxidized by bacteria to produce acetic acid [10]. The concentration and composition of the different organic acids determine the taste and aroma of the products [6]. Thus, in order to obtain a sensorial pleasant drink, according to literature, fermentation must end when the total acidity reaches the ideal range of 4 to 5 g / L [48]. In our study, the analysis of total titratable acidity indicated values between 1.80 ± 0.59 and 4.86 ± 0.72 g/L. In addition, the results of measuring total titratable acidity can be influenced by the addition of components in secondary fermentation, such as fruits and spices, which also have a very variable matrix [9,49,50]. According to the variation in the results obtained by evaluating the physicochemical parameters of kombucha samples in this work, a diversity of kombuchas is present in the Brazilian market. Since approximately 25 small companies currently market their products, the data generated by this study represent an initial mapping that has helped to create an improved understanding of the composition of kombuchas. Moreover, the use of simple methodologies for sample characterization, such as those used in this work, serves to indicate a method for fast and efficient quality control of this drink.
|
Physicochemical parameters |
||||
|
Brands |
Alcohol % (V/V) |
pH |
Total titratable acidity (g/L Hac) |
|
|
k1 |
2,54 ± 0,39 a,b,c (***) |
3,23±0,08 c (*) |
4,55±0,45 b (***), e (**) |
|
|
k2 |
1,11 ± 0,38 |
2,94±0,33 |
4,86±0,72 f (***), i (**), h (*) |
|
|
k3 |
1,07 ± 0,32 |
3,24±0,06 j (*) |
1,80±0,59 |
|
|
k4 |
1,03 ± 0,24 |
2,88±0,06 |
3,20±0,34 |
|
|
k5 |
2,02 ± 0,42 h,k,m (*) |
3,21±0,06 |
2,80±0,56 |
|
|
k6 |
1,85±0,35 n (*) |
3,43±0,04 i,o (*) n (***) |
2,52±0,60 |
|
|
Values expressed as mean ± OD. ANOVA test with Dunn's posteriori for the “Brands” variable. Values considered statistically significant with p *** <0.001. **p <0,01. *p <0,05. a) K1 vs. K2; b) K1 vs. K3; c) K1 vs. K4; d) K1 vs. K5; e) K1 vs. K6, f) K2 vs. K3; g) K2 vs. K4; h) K2 vs. K5; i) K2 vs. K6, j) K3 vs. K4; k) K3 vs. K5; l) K3 vs. K6; m) K4 vs. K5; n) K4 vs. K6; o) K5 vs. K6. |
||||
Table 1: Physicochemical parameters of six different kombucha brands
4. Conclusion
This study was the first to use HTS to analyse the microbiome of kombuchas produced and commercialized in Brazil. Here, the taxonomy of microorganisms found in six Brazilian kombucha brands was identified. However, their respective contributions to the fermentation process and the organoleptic characteristics of the kombucha have yet to be elucidated. In addition, the difference in physicochemical composition found between the brands can be explained by the use of different matrices in the preparation of kombuchas, different origins of SCOBY acquisition, in addition to fermentation and handling conditions. In this way, the data generated in this work stimulate new scientific discussions and investigations about kombucha and highlight the importance of carrying out analyses and adequate labelling. Finally, together with additional data from samples collected on the spot by the Ministry of Agriculture, Livestock and Supply (MAPA), the physicochemical data obtained in this study helped to build the standard of identity and quality of kombucha in Brazil (PIQ), resulting in Normative Instruction 41/2019 [47].
Funding
We thank the Conselho Nacional de Desenvolvimento Científico e Tecnológico do Brasil (CNPq) #305495/2018-6 and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES).
Acknowledgments
The authors kindly thank the owners of the companies that provided the kombuchas.
Conflicts of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Data availability (recommended)
The datasets generated for this study can be found in the Illumina reads generated in this study have been submitted to NCBI-SRA under Bio Project number PRNJA611694 in the NCBI BioProject database.
References
- The Kombucha Industry (2020).
- Tran T, Grandvalet C, Verdier F, et al. Microbiological and technological parameters impacting the chemical composition and sensory quality of kombucha. Compr Rev Food Sci Food Saf 19 (2020): 2050-2070.
- Brazilian Association of Kombucha. Associates. Our members (2020).
- Leal JM, Suarez LV, Jayabalan R, et al. A review on health benefits of kombucha nutritional compounds and metabolites. Cyta-Journal of Food 16 (2018): 390-399.
- Teoh AL, Heard G, Cox J. Yeast ecology of Kombucha fermentation. International Journal of Food Microbiology 95 (2004): 119-126.
- Chen C, Liu BY. Changes in major components of tea fungus metabolites during prolonged fermentation. Journal of Applied Microbiology 89(2000): 834-839.
- Martini N. Kombucha. Journal of Primary Health Care 10 (2018): 93-94.
- Coton M, Pawtowski A, Taminiau B. Unraveling microbial ecology of industrial-scale Kombucha fermentations by metabarcoding and culture-based methods. FEMS Microbiology Ecology 93(2017).
- Talebi M, Frink LA, Patil RA, et al. Examination of the Varied and Changing Ethanol Content of Commercial Kombucha Products. Food Analytical Methods 10(2017): 4062-4067.
- May A, Narayanan S, Alcock J, et al. Kombucha: a novel model system for cooperation and conflict in a complex multi-species microbial ecosystem. Peerj 7 (2019).
- Jayabalan R, Malbasa RV, Loncar ES, et al. A Review on Kombucha Tea-Microbiology, Composition, Fermentation, Beneficial Effects, Toxicity, and Tea Fungus. Compr Rev Food Sci Food Saf 13 (2014): 538-550.
- Villarreal-Soto SA, Beaufort S, Bouajila J, et al. Understanding Kombucha Tea Fermentation: A Review. Journal of Food Science 83 (2018): 580-588.
- Primiani CN, Pujiati, Mumtahanah M, et al. Kombucha fermentation test used for various types of herbal teas. 7th International Seminar on New Paradigm and Innovation on Natural Science and Its Application 1025 (2018).
- Watawana MI, Jayawardena N, Gunawardhana CB, et al. Health, Wellness, and Safety Aspects of the Consumption of Kombucha. Journal of Chemistry (2015).
- Caporaso JG, Lauber CL, Walters WA, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proceedings of the National Academy of Sciences of the United States of America 108 (2011): 4516-4522.
- White T, Bruns T, Lee S, et al. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics 31 (1990): 315-322.
- Escudié F, Auer L, Bernard M, et al. FROGS: Find, Rapidly, OTUs with Galaxy Solution. Bioinformatics, 34 (2018): 1287-1294.
- Rognes T, Flouri T, Nichols B, et al. VSEARCH: a versatile open source tool for metagenomics. Peerj 4 (2016).
- Kim M, Oh HS, Park SC, et al. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. International Journal of Systematic and Evolutionary Microbiology 64 (2014): 346-351.
- McMurdie PJ, Holmes S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLOS ONE 8 (2013): e61217.
- Ministério da Agricultura Pecuária e Abastecimento MAPA (Instrução normativa n°24, 8 setembro de 2005). Aprova o Manual Operacional de Bebidas e Vinagres (2005).
- De Filippis F, Parente E, Ercolini D. Recent Past, Present, and Future of the Food Microbiome. Annual Review of Food Science and Technology 9 (2018): 589-608.
- Marsh AJ, O'Sullivan O, Hill C, et al. Sequence-based analysis of the bacterial and fungal compositions of multiple kombucha (tea fungus) samples. Food Microbiology 38 (2014): 171-178.
- Bevardi M, Frece J, Mesarek D,et al. Antifungal and Antipatulin Activity of Gluconobacter Oxydans Isolated from Apple Surface. Arhiv za higijenu rada i toksikologiju 64 (2013): 93-98.
- Lorentzen M, Campbell-Sills H, Jorgensen TS, et al. Expanding the biodiversity of Oenococcus oeni through comparative genomics of apple cider and kombucha strains. BMC Genomics 20 (2019): 330.
- Lorentzen M, Lucas P. Distribution of Oenococcus oeni populations in natural habitats. Applied Microbiology and Biotechnology 103 (2019): 2937-2945.
- Savary O, Mounier J, Thierry A, et al. Tailor-made microbial consortium for Kombucha fermentation: Microbiota-induced biochemical changes and biofilm formation. Food Research International 147 (2021): 110549.
- Edwards CG, Collins MD, Lawson PA, et al. Lactobacillus nagelii sp. nov., an organism isolated from a partially fermented wine. International Journal of Systematic and Evolutionary Microbiology, 50 (2000): 699-702.
- Nielsen D, Teniola O, Ban-Koffi L, et al. The microbiology of Ghanaian cocoa fermentations analysed using culture-dependent and culture-independent methods. International Journal of Food Microbiology 114 (2007): 168-186.
- Fabricio MF, Mann MB, Kothe CI, et al. Effect of freeze-dried kombucha culture on microbial composition and assessment of metabolic dynamics during fermentation. Food Microbiology 101 (2022): 103889.
- Gaggìa F, Baffoni L, Galiano M, et al. Kombucha Beverage from Green, Black and Rooibos Teas: A Comparative Study Looking at Microbiology, Chemistry and Antioxidant Activity. Nutrients 11 (2018): 1.
- Sreeramulu G, Zhu Y, Knol W. Kombucha Fermentation and Its Antimicrobial Activity. Journal of Agricultural and Food Chemistry 48 (2000): 2589-2594.
- Machado RTA, Gutierrez J, Tercjak A, et al. Komagataeibacter rhaeticus as an alternative bacteria for cellulose production. Carbohydrate Polymers 152 (2016): 841-849.
- Semjonovs P, Ruklisha M, Paegle L, et al. Cellulose synthesis by Komagataeibacter rhaeticus strain P 1463 isolated from Kombucha. Applied Microbiology and Biotechnology 101 (2017): 1003-1012.
- Gupta A, Singh VK, Qazi G, et al. Gluconobacter oxydans: Its biotechnological applications. Journal of molecular microbiology and biotechnology 3 (2001): 445-456.
- Schifferdecker A, Dashko S, Ishchuk O, et al. The wine and beer yeast Dekkera bruxellensis. Yeast (Chichester, England), 31 (2014).
- Mayser P, Fromme S, Leitzmann G, et al. The yeast spectrum of the ‘tea fungus Kombucha’. Mycoses, 38 (1995): 289-295.
- Renouf V, Falcou M, Miot-Sertier C, et al. Interactions between Brettanomyces bruxellensis and other yeast species in the initial stages of winemaking. Journal of Applied Microbiology 100 (2006): 1208-1219.
- Bellut K, Michel M, Hutzler M, et al. Investigation into the Potential of Lachancea fermentati Strain KBI 12.1 for Low Alcohol Beer Brewing. Journal of the American Society of Brewing Chemists 77 (2019): 157-169.
- Comitini F, Gobbi M, Domizio P, et al. Selected non-Saccharomyces wine yeasts in controlled multistarter fermentations with Saccharomyces cerevisiae. Food Microbiology 28 (2011): 873-882.
- Porter TJ, Divol B, Setati ME. Lachancea yeast species: Origin, biochemical characteristics and oenological significance. Food Research International 119 (2019): 378-389.
- Leuck AM, Rothenberger MK, Green JS. Fungemia due to Lachancea fermentati: a case report. BMC Infectious Diseases 14 (2014): 250.
- Clemente-Jimenez JM, Mingorance-Cazorla L, Martinez-Rodriguez S, et al. Molecular characterization and oenological properties of wine yeasts isolated during spontaneous fermentation of six varieties of grape must. Food Microbiology 21 (2004): 149-155.
- Amarasinghe H, Weerakkody NS, Waisundara VY. Evaluation of physicochemical properties and antioxidant activities of kombucha “Tea Fungus” during extended periods of fermentation. Food Science & Nutrition 6 (2018): 659-665.
- Nummer B. Kombucha Brewing Under the Food and Drug Administration Model Food Code: Risk Analysis and Processing Guidance. Journal of environmental health 76 (2013): 8-11.
- Neffe-Skocinska K, Sionek B, Scibisz I, et al. Acid contents and the effect of fermentation condition of Kombucha tea beverages on physicochemical, microbiological and sensory properties. CyTA - Journal of Food 15 (2017): 601-607.
- Ministry of Agriculture, Livestock and Food Supply MAPA. Instrução normativa n°41, de 17 de setembro de (2019).
- Velicanski AS, Cvetkovic DD, Markov SL, et al. Antioxidant and Antibacterial Activity of the Beverage Obtained by Fermentation of Sweetened Lemon Balm
 (Melissa officinalis L.) Tea with Symbiotic Consortium
(Melissa officinalis L.) Tea with Symbiotic Consortium  of Bacteria and Yeasts. Food technology and biotechnology, 52 (2014): 420-429.
of Bacteria and Yeasts. Food technology and biotechnology, 52 (2014): 420-429. - Reva ON, Zaets IE, Ovcharenko LP, et al. Metabarcoding of the kombucha microbial community grown in different microenvironments. AMB Express 5 (2015): 35.
- Mahe F, Rognes T, Quince C, et al. Swarm: robust and fast clustering method for amplicon-based studies. Peerj 2 (2014).
Supplemental Information
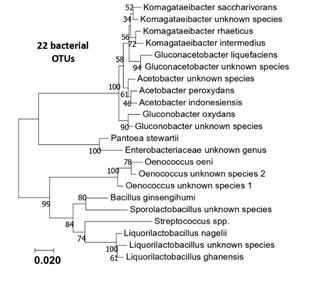
Figure S1: Phylogenetic tree of the 22 bacterial OTUS (16S rRNA sequences) generated from the six kombucha samples.
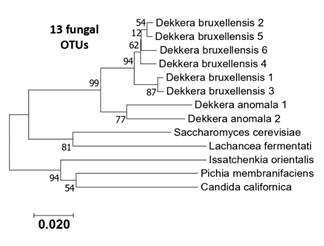
Figure S2: Phylogenetic tree of the 13 eukaryotic OTUS (ITS sequences) generated from the six kombucha samples.


 Impact Factor: * 3.8
Impact Factor: * 3.8 Acceptance Rate: 77.96%
Acceptance Rate: 77.96%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 