Boiling Extraction Method VS Commercial Kits for Bacterial DNA Isolation from Food Samples
Maria-Eleni Dimitrakopoulou, Venia Stavrou, Chrysoula Kotsalou, Apostolos Vantarakis*
Department of Public Health, Medical School, University of Patras, Patras, Greece
*Corresponding Author: Apostolos Vantarakis, Department of Public Health, Medical School, University of Patras, Patras, Greece
Received: 25 November 2020; Accepted: 10 December 2020; Published: 21 December 2020
Article Information
Citation:
Maria-Eleni Dimitrakopoulou, Venia Stavrou, Chrysoula Kotsalou, Apostolos Vantarakis. Boiling extraction method VS commercial Kits for bacterial DNA isolation from food samples. Journal of Food Science and Nutrition Research 3 (2020): 311-319.
DOI: 10.26502/jfsnr.2642-11000057
View / Download Pdf Share at FacebookAbstract
Background: In recent years, the development of commercial Kits for DNA extraction, combined with the use of molecular technologies have been utilized for the understanding of microbial ecosystems. DNA extraction considered being a critical step for molecular techniques such as Polymerase Chain Reaction. Several different methods and commercial kits for this purpose are nowadays available. The aim of this study was to compare five commercially available kits and, a cheap, rapid, non-commercial method, the boiling method for isolating bacterial DNA from different food matrixes.
Methods: Experimental protocol was applied on four different types of food: olives, roe, raisins and bacon. The evaluation of the results was performed first by using a spectrophotometer, by measuring purity and concentration of the extracted DNA, followed by electrophoresis where the integrity of the DNA was checked and finally by PCR-amplification.
Results: Results revealed that boiling method is efficient, simple, cheap and suitable for PCR amplification for all these food types.
Conclusion: Boiling method for bacterial DNA extraction from food should be further explored in order to be validated and established.
Keywords
<p>Bacterial DNA extraction, Food sample, Boiling method, Polymerase Chain Reaction, 16rRNAgene</p>
Article Details
1. Introduction
Until recently, the knowledge and understanding of research community on microbial ecosystems was based mostly on cultivation methods. Due to the need for direct results, DNA-based methods that are culture-independent have been developed in recent years. These methods offer the ability to detect microorganisms in food, as well as to identify their microbial communities [1, 2]. DNA-based methods are characterized by high specificity, reproducibility, sensitivity, speed and are also cost effective [3]. These methods, however, depend on the DNA extracted protocols from a sample. Food sample considered as being a difficult matrix due to the presence of inhibitors such as fats, proteins, and calcium. Moreover, incomplete cell lysis, degraded or damaged extracted DNA and adsorption of DNA into a specific material or simultaneous extraction together with enzyme inhibitors are the most common problems of DNA extraction process [4-6]. Consequently, it is clear that the extraction of high purity intact bacterial DNA, at high concentrations, from food samples is a demanding process and constitutes a critical step in molecular techniques [7]. The first step in the challenging process of DNA extraction is cell lysis. Especially for food microbiota, the rupture of the cell membrane, following the inactivation of cellular nucleases and the separation of the desired nucleic acid from cellular debris, is a hurdle [8]. Several methods have been already applied for bacterial cell wall lysis and DNA extraction based on detergents, proteolytic enzymes, lysozyme, mechanical disruption or temperature changes [9]. Phenol chloroform method, or the combination of chemical and physical methods are mostly used, but are laborious, non-cost-effective and include handling of highly toxic reagents [10]. Recently, commercial kits for DNA extraction were developed. Although they are simpler DNA extraction protocols that are not so laborious, but they also constitute an expensive choice [11]. Due to their high cost especially for developing countries, and the continuous need for accredited methods, international standardization requires in-house methods to be established [12]. Boiling considered being a suitable method for DNA extraction of a variety of microorganisms. This method is already largely used worldwide and characterized for its simple protocol and low cost [13]. As far as food samples concerns, physical, chemical and biological characteristics of each matrix can affect purity and concentration of extracted DNA by this method [14]. In the present study, we evaluated the extraction of bacterial DNA from four different food samples using direct boiling method and compared the results with five different commercial kits. Fresh produce, dry fruits, sea and animal products are food types with a great variety of bacteria communities and are also considered to be complex matrices [15-18]. Therefore, olives, raisins, fish eggs and bacon were chosen as matrices. The main point of this research is the comparison of the quality of the bacterial extracted DNA with available commercial isolation kits to the boiling method and the determination of efficiency, simplicity, cost and duration of each method.
2. Materials and Methods
2.1 Food Samples
We compared the effect of several matrices on the bacterial DNA extraction process. Fresh olive samples were collected, under aseptic conditions, right after harvest and transferred to laboratory. Raisins were also collected under aseptic conditions after the drying process. Moreover, roe (fish eggs) and bacon are used as food samples too and purchased from local super market. All food samples were collected in triplicates and store at -20°C until analysis.
2.2 DNA extraction methods
Six DNA extraction methods were evaluated to compare their relative efficiency with respect to the extraction of DNA from food samples. The characteristics of the DNA extraction methods are summarized in Table 1. All commercial kits were used according to the protocol described by manufacturers. According to manufacturers of Nucleospin Tissue and QiAmp DNA Stool Mini Kit, indicated that Lysis Buffer T1 and ASL were high-strength chaotropic guanidium salts and detergents, with washing buffers consisting of initial low strength chaotropic salts, and Tris/alcohol/acid buffers for DNA elution. According to boiling method protocol, 10 g of food were homogenized with 90 ml of Brain Heart Infusion in a stomacher. 10 ml were transferred to a falcon and placed for incubation at 30°C for 18 hours. This was followed by centrifuge at 14,000 g for 20 minutes. The precipitate was transferred to an Eppendorf with 1 ml of sterile water and placed in a water bath at 100°C for 10 minutes [19]. The centrifuge was repeated at 1000 g for 5 minutes. The supernatant was stored at -20°C. Five replicates were performed for each method per food sample.
|
Method |
Technology |
Source |
Cell lysis |
|
Nucleospin Plant II |
Silica-membrane |
Commercial |
CTAB lysis/SDS lysis method |
|
Nucleospin Tissue |
Silica-membrane |
Commercial |
Lysis Buffer TI |
|
QiAmp DNA Stool Mini kit |
Silica-membrane |
Commercial |
Lysis Buffer ASL |
|
DNeasy Ultraclean Microbial kit |
Silica-membrane |
Commercial |
Combination of heat, detergent and mechanical force |
|
Dneasy Mericon Food kit |
Silica-membrane |
Commercial |
Modified CTAB lysis method |
|
Boiling |
Overheat 100C |
Non-Commercial |
Cell lysis with boiling |
Table 1: Characteristics of extraction methods.
2.3 Quantification and quality assessment of DNA extraction products
The isolated DNA was analyzed by 0,8%(w/v) agarose gel electrophoresis using 1000bs ladder as marker. Purity and concentration of DNA were estimated by Nanodrop™ 1000 spectrophotometer. Yield of extracted DNA was determined by measuring absorbance at 260 nm. The purity of corresponding DNA samples was determined by A260/A280 (DNA/protein) and A260/A230 (DNA/humic acid) ratios to determine protein and humic acid contamination, respectively.
2.4 PCR amplification of the bacterial community 16S rRNA gene
The intact DNA extracts were used as a template for PCR amplification of 16s rRNA (V3 region, 240bs) using universal 16s primers. The V3 region of each sample was amplified with primer 338F (5′ACTCCTACGGGGGCAGCAG, Sigma, France) and518R (5′ATTACCGCGGCTGCTGG, Sigma, France) [20-22]. The PCR reaction was carried out in a final volume of 25μl, containing 100mg DNA template, 5μL 5buffer C Mg free, 1,5μL MgCl2 (25mM), 0,5μL dNTPs (10mM), 5μL primers (1μΜ) and 0,1μL Taq polymerase (Kapa TAq PCR kit) (Sigma, France). PCR amplification was performed using Thermocycler (Biorad). The amplification program was initial denaturation 95°C for 3 mins, followed by 30 cycles of denaturing 95°C for 1 min, annealing at 55°C for 1 min, extension at 72°C for 1min with a final extension at 72°C for 10 mins. Aliquots of PCR products analyzed by electrophoresis in 2% (w/v) agarose gel. Electrophoresis was performed using 1× Tris–acetic acid and EDTA (TAE) buffer, stained with 8μL of GelRed (Biotium) and run at a constant voltage of 80 V for 120 min. The DNA bands were visualized and images were acquired using UVP.
3. Results
3.1 Time and cost performance of DNA extraction
The cost of analysis per sample for non-commercial method was based on the cost of the reagents and solutions used. For commercial methods, cost based on the price of each kit in Greece. Moreover, time, dedicated to the analysis for one sample, was defined. The boiling method, as shown in Table 2, is the cheapest and requires the less time to be performed.
3.2 Visualizing DNA by agarose gel electrophoresis
In the present study, 2% agarose concentration for electrophoresis was used.
3.3 Comparison of the yield and purity of bacteria DNA extracted using a variety of extraction methods
A 260/280 ratio in this study was found to be in a range of 0.7-2.0. Table 3 summarizes the DNA yield and purity range obtained for all bacteria extracts using the six extraction methods. The average values of measures obtained by Nanodrop from initially extracted gDNA samples are shown in Table 3. Results revealed that Boiling Method gave the highest genomic DNA yield in all four food matrixes. From olives, roe and bacon, Dneasy Ultraclean Microbial Kit generated the lowest genomic DNA yield. In contrast, the lowest genomic DNA yield from raisins obtained from QiAmp DNA Stool Mini Kit.
|
Method |
Cost/sample € |
Avergae Time |
|
Nucleospin Plant II |
4,3 |
30 mins |
|
Nucleospin Tissue |
3,96 |
> 1h |
|
QiAmp DNA Stool Mini kit |
5,7 |
50 mins |
|
DNeasy Ultraclean Microbial kit |
4,3 |
50 mins |
|
Dneasy Mericon Food kit |
5,4 |
1h |
|
Boiling |
0 |
20 mins |
Table 2: Price and Time cost each method per sample.
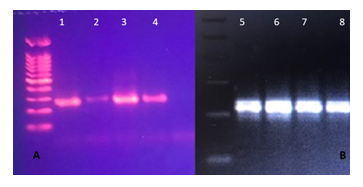
Figure 1: Agarose gel electrophoresis of bacteria DNA. (A) Dneasy Mericon food kit from bacon and raisins (1,2) QiAmp DNA stool Mini Kit from roe and olives (3, 4); (B) Boiling method for bacteria DNA extraction from olives, roe, raisins, bacon (5, 6, 7, 8).
|
Food Sample |
DNA Extraction Method |
DNA yield ng/ml-1 |
DNA Purity 260/280 |
16s PCR |
|
Olive |
Nucleo spin Plant II |
17,9 |
1,3 |
+ |
|
Olive |
Nucleo spin Tissue |
28,7 |
1,59 |
|
|
Olive |
QiAmp DNA Stool Mini kit |
7,9 |
1,55 |
+ |
|
Olive |
DNeasy Ultraclean Microbial kit |
5,2 |
1,3 |
|
|
Olive |
Dneasy Mericon Food kit |
7,2 |
1,5 |
|
|
Olive |
Boiling |
622,11 |
1,8 |
+ |
|
Roe |
Nucleospin Plant II |
1492,66 |
1,9 |
|
|
Roe |
Nucleospin Tissue |
102,25 |
1,2 |
+ |
|
Roe |
QiAmp DNA Stool Mini kit |
571,5 |
2 |
+ |
|
Roe |
DNeasy Ultraclean Microbial kit |
3,46 |
1,21 |
|
|
Roe |
Dneasy Mericon Food kit |
15,2 |
1,6 |
|
|
Roe |
Boiling |
862,2 |
1,8 |
+ |
|
Raisins |
Nucleospin Plant II |
12,55 |
0,76 |
+ |
|
Raisins |
Nucleospin Tissue |
32 |
1,3 |
|
|
Raisins |
QiAmp DNA Stool Mini kit |
1,5 |
1,6 |
|
|
Raisins |
DNeasy Ultraclean Microbial kit |
5,9 |
1,4 |
|
|
Raisins |
Dneasy Mericon Food kit |
2,25 |
1,3 |
+ |
|
Raisins |
Boiling |
565,1 |
1,6 |
+ |
|
Bacon |
Nucleospin Plant II |
10,2 |
1,5 |
|
|
Bacon |
Nucleospin Tissue |
7,9 |
1,4 |
|
|
Bacon |
QiAmp DNA Stool Mini kit |
17,8 |
1,4 |
+ |
|
Bacon |
DNeasy Ultraclean Microbial kit |
4,9 |
1,5 |
|
|
Bacon |
Dneasy Mericon Food kit |
50,51 |
1,9 |
+ |
|
Bacon |
Boiling |
211,1 |
1,9 |
+ |
Table 3: Comparison of DNA purity and yield obtained by six different methods. Presence of amplicon after PCR amplification.
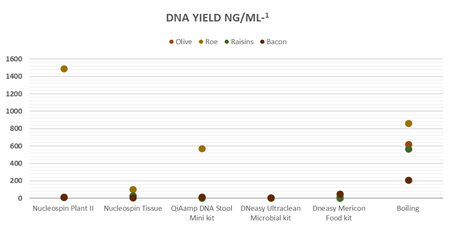
Figure 2: DNA yield extracted from four food types each method.
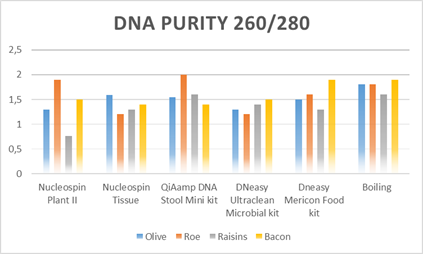
Figure 3: DNA purity per sample and method.
4. Discussion
DNA based molecular analysis of a food sample requires the efficient extraction of DNA of the sample. In the present study, five commercial methods and one non-commercial method were assessed to compare their relative success with respect to the bacterial DNA extraction from four different food samples. More specific, these methods were examined to assess their relative ability to extract bacterial DNA at a high concentration and purity and facilitate subsequent PCRs. Moreover, these six methods for bacterial DNA extraction were compared with each other regarding to cost and average time needed per sample.
There are numerous studies that have used these commercial kits for bacterial DNA extraction, despite the fact that they are not designed for that purpose [23-25]. Nucleospin Plant II designed for efficient extraction of genomic DNA from plant tissue using CTAB or SDS lysis buffer. Manufacturers recommend homogenization of samples with mortar and pestle in presence of liquid nitrogen. However, other homogenization such as rotor-stator homogenizer or bead mills can be suitable. In the present study, mortar and pestle in presence of liquid nitrogen are used. Nucleospin Tissue is recommended to be used for purification of total DNA from clinical or forensic samples, tissues, cells, yeast, bacteria, or viruses. QiAmp DNA Stool Mini kit is designed for purification of genomic, bacterial, viral, and parasite DNA from fresh or frozen human stool or other sample types with high concentrations of PCR inhibitors. According to manufactures, Dneasy Ultraclean Microbial Kit, is able to isolate high quality genomic DNA from 1,8ml microbial culture, including bacterial and fungal spores. Dneasy Mericon Food Kit designed for DNA extraction from raw and processed foods. Homogenization in presence of liquid nitrogen is recommended. The need of liquid nitrogen in NucleoSpin Plant II and Dneasy Mericon Food Kit is a restrictive factor for lab routine analysis and thus can be substituted.
Although, these commercial kits were not specifically designed for bacterial DNA extraction from food samples, some of them, depending on food matrix, can be suitable for this purpose.
In contrast, boiling method gave satisfactory results for every food type examined, with successful purification. More specifically, it is observed that the use of boiling method is an effective method for the isolation of bacterial DNA from olives and raisins. As for the roe, the DNA extraction with the QiAamp DNA Stool Mini kit proved to be as effective as boiling. Despite the high concentration of DNA and its purity with the use of Nucleospin Plant II we conclude from the electrophoresis that the DNA is degraded. The use of boiling method and the Dneasy Mericon Food kit were the most effective methods for isolating bacterial DNA from bacon. Boiling protocol though, starts with 10 g from food sample in contrast to commercial kits which use 2 g from food.
As far as duration time for each method is concerned, boiling method is simple, rapid and doesn’t need high experience. It is also the most cost-effective method in contrast to commercial kits, which prices range between 200-300 € for 50 reps. The use of commercials kits need some experience, due to many material handling, and demand special conditions for storage comparing to boiling method.
5. Conclusion
In this study, five commercial kits and a non-commercial method for bacterial DNA extraction from food samples were compared. Boiling method proved to be the most efficient for the isolation of bacterial DNA from all 4 different types of food in terms of purity and concentration. Furthermore, the presence of an amplification band in all the different samples after PCR by the boiling method documents the effectiveness of the method in extracting bacterial DNA from the specific matrixes. We concluded that boiling method is a simple, economical, fast method without the need for specialized reagents and is an appropriate alternative for carrying out molecular studies, compared to the commercial Kits tested in the four specific food matrices. Since, boiling method seems to be an efficient way to extract bacterial DNA from these samples we can assume that the same DNA is suitable for other molecular techniques, toο. Some studies confirming that our hypothesis already exists and boiling method has applied in a great variety of molecular methods, such as NGS, PCR, qPCR, PCR-DGGE, etc. [26-29]. Nevertheless, further studies are necessary in order to evaluate the method for more matrixes and other molecular techniques.
Concluding, our study, brings the scientific community one step closer to the in house method that Yalç?nkaya et al. [12] are looking for and boiling method seem to be a promising method. More studies are necessary to ensure the specificity, reproducibility, and sensitivity of the boiling method.
Acknowledgments
The present research received fund from the Single State Action Aid for Research, Technological Development & Innovation “INVESTIGATE - CREATE - INNOVATE” project “Trust Trace” T1EDK-04028.
References
- Ceuppens S, Li D, Uyttendaele M, et al. Molecular methods in food safety microbiology: Interpretation and implications of nucleic acid detection. Compr Rev Food Sci Food Saf 13 (2014): 551-577.
- Mateus-Barros E, Meneghine AK, Bagatini IL, et al. Comparison of two DNA extraction methods widely used in aquatic microbial ecology. J Microbiol Methods 159 (2019): 12-17.
- Pinto A Di, Forte VT, Guastadisegni MC, et al. A comparison of DNA extraction methods for food analysis. Food Control 18 (2007): 76-80.
- Di Bernardo G, Del Gaudio S, Galderisi U, et al. Comparative evaluation of different DNA extraction procedures from food samples. Biotechnol Prog 23 (2007): 297-301.
- Jara C, Mateo E, Guillamón JM, et al. Analysis of several methods for the extraction of high quality DNA from acetic acid bacteria in wine and vinegar for characterization by PCR-based methods. Int J Food Microbiol 128 (2008): 336-341.
- Kuhn R, Böllmann J, Krahl K, et al. Comparison of ten different DNA extraction procedures with respect to their suitability for environmental samples. J Microbiol Methods 143 (2017): 78-86.
- Quigley L, O’Sullivan O, Beresford TP, et al. A comparison of methods used to extract bacterial DNA from raw milk and raw milk cheese. J Appl Microbiol 113 (2012): 96-105.
- Abdelhai M. Comparative Study of Rapid DNA Extraction Methods of Pathogenic Bacteria. Am J Biosci Bioeng 4 (2016): 1.
- Hannaa F, Ibtisam H. Evaluation three methods of the extraction and purification of bacterial DNA of Gram positive and Gram negative bacteria. World J Exp Biosci 4 (2016): 62-65.
- Javadi A, Shamaei M, Ziazi LM, et al. Qualification study of two genomic DNA extraction methods in different clinical samples. Tanaffos 13 (2014): 41-47.
- Norliza Nik Hassan N, Hanis Zainal Abidin N, Alisya Ahmad Saifuddin A, et al. Efficacy of DNA Extraction Methods Using Simple Boiling and Commercial Kit in Extracting Streptococcus Mutans in Caries. Malaysian J Hum Genet 1 (2020): 26-34.
- Yalç?nkaya B, Yumbul E, Mozio?lu E, et al. Comparison of DNA extraction methods for meat analysis. Food Chem 221 (2017): 1253-1257.
- Barbosa C, Nogueira S, Gadanho M, et al. DNA Extraction: Finding the Most Suitable Method. Elsevier Inc (2016).
- Giacomazzi S, Leroi F, Joffraud JJ. Comparison of three methods of DNA extraction from cold-smoked salmon and impact of physical treatments. J Appl Microbiol 98 (2005): 1230-1238.
- Argyri K, Doulgeraki AI, Manthou E, et al. Microbial diversity of fermented greek table olives of halkidiki and konservolia varieties from different regions as revealed by metagenomic analysis. Microorganisms 8 (2020): 1-18.
- Zhao X, Han L, Xiao J, et al. Jo l P. Sci Total Environ (2019).
- Yi L, Su G, Hu G, et al. Diversity study of microbial community in bacon using metagenomic analysis. J Food Saf 37 (2017): 1-9.
- Wei YJ, Wu Y, Yan YZ, et al. High-throughput sequencing of microbial community diversity in soil, grapes, leaves, grape juice and wine of grapevine from China. PLoS One 13 (2018): 1-17.
- Ribeiro JC, Tamanini R, Soares BF, et al. Efficiency of boiling and four other methods for genomic DNA extraction of deteriorating spore-forming bacteria from milk. Semin Agrar 37 (2016): 3069-3078.
- Salam M, Varma A. Bacterial community structure in soils contaminated with electronic waste pollutants from Delhi NCR, India. Electron J Biotechnol 41 (2019): 72-80.
- Ding C, Adrian L, Peng Y, et al. 16S rRNA gene-based primer pair showed high specificity and quantification accuracy in detecting freshwater Brocadiales anammox bacteria. FEMS Microbiol Ecol 96 (2019): 1-11.
- Amorim CL, Alves M, Castro PML, et al. Bacterial community dynamics within an aerobic granular sludge reactor treating wastewater loaded with pharmaceuticals. Ecotoxicol Environ Saf 147 (2018): 905-912.
- Smith B. Optimising Bacterial DNA Extraction from Faecal Samples: Comparison of Three Methods. Open Microbiol J 5 (2011): 14-17.
- Chaillou S, Chaulot-Talmon A, Caekebeke H, et al. Origin and ecological selection of core and food-specific bacterial communities associated with meat and seafood spoilage. ISME J 9 (2015): 1105-1118.
- Riquelme C, Câmara S, Enes Dapkevicius M de LN, et al. Characterization of the bacterial biodiversity in Pico cheese (an artisanal Azorean food). Int J Food Microbiol 192 (2015): 86-94.
- Xiong X, Huang M, Yuan F, et al. Development and Validation of a Fast DNA Extraction Protocol for Fish Products. Food Anal Methods 12 (2019): 1998-2008.
- Yamagishi J, Sato Y, Shinozaki N, et al. Comparison of boiling and robotics automation method in DNA extraction for metagenomic sequencing of human oral microbes. PLoS One 11 (2016): 1-15.
- Dilhari A, Sampath A, Gunasekara C, et al. Evaluation of the impact of six different DNA extraction methods for the representation of the microbial community associated with human chronic wound infections using a gel-based DNA profiling method. AMB Express 7 (2017).
- Mohd Nosi MZ, Syed Jamil Fadaak SNE, Muhammad MDD, et al. Assessment of gut microbiota in different developmental stages of Malaysian Mahseer (Tor tambroides). Aquac Res 49 (2018): 2977-2987.


 Impact Factor: * 3.8
Impact Factor: * 3.8 Acceptance Rate: 77.96%
Acceptance Rate: 77.96%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 