Impact of RAS-Pathway Activation on Phenotype and Outcome in Patients with Chronic Myelomonocytic Leukemia and a TET2/SRSF2 Comutation
Klaus Geissler1,2*, Eva Jäger3, Agnes Barna4, Michael Gurbisz3, Temeida Graf2, Elmir Graf2, Maike Stegemann2, Thomas Nösslinger5, Michael Pfeilstöcker5, Sigrid Machherndl-Spandl6, Reinhard Stauder7, Armin Zebisch8,9, Heinz Sill8, Leopold Öhler10, Rajko Kusec11, Gregor Hoermann3,12,13, and Peter Valent13,14
1Medical School, Sigmund Freud University, Vienna, Austria
2Department of Internal Medicine V with Hematology, Oncology and Palliative Medicine, Hospital Hietzing, Vienna, Austria
3Department of Laboratory Medicine, Medical University of Vienna, Vienna, Austria
4Blood Transfusion Service, Blood Transfusion Service for Upper Austria, Austrian Red Cross, Linz, Austria
5Department of Internal Medicine III, Hanusch Hospital, Vienna, Austria
6Department of Internal Medicine I with Hematology with Stem Cell Transplantation, Hemostaseology and Medical Oncology, Ordensklinikum Linz Barmherzige Schwestern - Elisabethinen, Linz, Austria
7Internal Medicine V with Hematology and Oncology, Medical University of Innsbruck, Innsbruck, Austria
8Department of Internal Medicine, Division of Hematology, Medical University of Graz, Graz, Austria
9Otto-Loewi Research Center for Vascular Biology, Immunology and Inflammation, Division of Pharmacology, Medical University of Graz, Austria
10Department of Internal Medicine/Oncology, St. Josef Hospital, Vienna, Austria
11School of Medicine, University of Zagreb, University Hospital Dubrava, Zagreb, Croatia
12Central Institute of Medical and Chemical Laboratory Diagnostics, Medical University of Innsbruck, Innsbruck, Austria
13Ludwig Boltzmann Institute for Hematology and Oncology (LBI HO), Medical University of Vienna, Vienna, Austria
14Department of Internal Medicine I, Division of Hematology and Hemostaseology, Medical University of Vienna, Vienna, Austria
*Corresponding Author: Dr. Klaus Geissler, Medical School, Sigmund Freud University, Vienna, and Department of Internal Medicine V with Hematology, Oncology and Palliative Medicine, Hospital Hietzing, Wolkersbergenstrasse 1, 1130 Vienna; Austria
Received: 25 November 2020; Accepted: 08 January 2021; Published: 18 January 2021
Article Information
Citation:
Klaus Geissler, Eva Jäger, Agnes Barna, Michael Gurbisz, Temeida Graf, Elmir Graf, Maike Stegemann, Thomas Nösslinger, Michael Pfeilstöcker, Sigrid Machherndl-Spandl, Reinhard Stauder, Armin Zebisch, Heinz Sill, Leopold Öhler, Rajko Kusec, Gregor Hoermann, Peter Valent. Impact of RAS-Pathway Activation on Phenotype and Outcome in Patients with Chronic Myelomonocytic Leukemia and a TET2/SRSF2 Comutation. Journal of Cancer Science and Clinical Therapeutics 5 (2021): 083-093.
View / Download Pdf Share at FacebookAbstract
In unselected patients with chronic myelomonocytic leukemia (CMML) the prognostic impact of RAS mutations remains unclear. Restricting analysis to molecularly defined subgroups such as to patients with a TET2/SRSF2 comutation may be more appropriate to study the effects of RAS pathway mutations on the phenotype and clinical outcome. 87/291 CMML patients in our unselected real world “Austrian Biodatabase for CMML” (ABCMML) were found to have a TET2/SRSF2 comutation. In 37 of them mutations in at least one RAS-pathway component (NRAS, KRAS, CBL, NF1 and PTPN11) were detected. Patients with additional RASopathy gene mutations had higher WBC counts, and lower Hb and platelet values. Moreover, RAS-pathway activation was more frequently associated with splenomegaly, signs of transformation and high growth factor independent colony growth. The median survival of TET2/SRSF2-mutant CMML patients with additional molecular RAS-pathway aberrations was 15 months as compared to 38 months in patients without such mutations (p=0.0026). Interestingly, the presence of RASopathy gene mutations was not associated with a poor outcome in CMML patients without a TET2/SRSF2 comutation. We confirm here in a common molecularly defined subgroup of CMML patients that RAS-pathway activation is associated with changes in the clinicohematologic phenotype and an inferior outcome.
Keywords
<p>CMML; in vitro cultures; CFU-GM; RAS; TET2; SRSF2; Prognosis</p>
Article Details
1. Introduction
Chronic myelomonocytic leukemia (CMML) is a molecularly and phenotypically complex and heterogenous disease [1-3]. Over the past few years, a large number of mutations in genes encoding epigenetic regulators (TET2, ASXL1, EZH2, UTX, IDH1, IDH2, DNMT3A), splicing factors (SF3B1, SRSF2, ZRSF2, U2AF1), and signaling molecules (NRAS, KRAS, CBL, JAK2, FLT3) have been identified in clonal cells in CMML [4]. In unselected patients with CMML the prognostic impact of RAS mutations remains unclear, with some studies, but not all, demonstrating inferior outcomes [5-8]. This fact may be due to variability of the mutational landscapes in patient populations because in molecularly heterogenous cancers important pathogenetic relationships may be masked in the molecular complexity of the disease. One way to uncover such relationships is the use of strictly molecularly defined subgroups to analyse the potential impact of additional molecular aberrations on the phenotype of disease and its clinical outcome. The combination of TET2 and SRSF2 mutations is very frequently observed in CMML and highly specific for myeloid neoplasm with monocytosis [2]. Therefore we chose this subgroup to analyse the impact of RAS-pathway mutations in this common molecularly defined category of CMML patients.
2. Methods
2.1 Patients
In the “Austrian Biodatabase for Chronic Myelomonocytic Leukemia” (ABCMML) clinicolaboratory real-life data have been captured from 606 CMML patients from 14 different hospitals over the last 30 years [9]. The ABCMML has been shown to be a representative and useful real-life data source for further biomedical research. 87 of 291 molecularly defined CMML patients in the ABCMML were found to have a TET2/SRSF2 comutation. In 37 of these patients additional mutations in at least one component of the RAS signaling pathway including NRAS, KRAS, CBL, NF1 and PTPN11, respectively, were detected.
2.2 Colony Assay
In one of our centers (Medical University of Vienna) the assessment of hematopoietic colony formation in vitro has been an integral part of the diagnostic work up in patients with suspected myeloid malignancies for many years [10]. Colony forming-unit granulocyte/macrophage (CFU-GM) growth was assessed in semisolid cultures without growth factors as previously described in one central laboratory [11]. Mononuclear cells (MNC) were isolated from PB of patients by Ficoll-Hypaque density gradient centrifugation (density 1.077 g/mL, 400g for 40 minutes). The low-density cells were collected from the interface between density solution and plasma, washed twice, and resuspended in Iscove‘s modified Dulbecco‘s medium (GIBCO, Paisley, Scotland). In unstimulated cultures PBMNCs were cultured in 0.9% methylcellulose, 30% fetal calf serum (FCS; INLIFE, Wiener Neudorf, Austria), 10% bovine serum albumin (Behring, Marburg, Germany), α-thioglycerol (10-4 mol/L) and Iscove‘s modified Dulbecco‘s medium. Cultures were plated in duplicates or triplicates, respectively, at 25-100 × 103 PBMNC/mL. In some cases the numbers of MNC chosen in our experiments were based on the colony growth in prior cell cultures in the respective patient in order to optimize evaluation of CFU-GM formation. Plates were incubated at 37°C, 5% CO2, and full humidity. After a culture period of 14 days, cultures were examined under an inverted microscope. Aggregates with more than 40 translucent, dispersed cells were counted as CFU-GM. CFU-GM data are expressed as mean values from cultures.
2.3 Molecular Studies
Genomic DNA was isolated from mononuclear cell (MNC) fractions of these blood samples according to standard procedures. The mutational status of CMML-related protein coding genes was determined by targeted amplicon sequencing using the MiSeq platform (Illumina). Details regarding gene panel, library preparation and data processing have been reported previously [9]. Only variants with an allelic frequency (VAF) ≥ 5%, a described population frequency (MAF) <1%, and an annotated pathogenic effect (or probability >90% of being pathogenic) were included, with pathogenicity determined according to databases as shown in Table S1 and published studies.
2.4 Statistical Analysis
The log-rank test was used to determine whether individual parameters were associated with OS. OS was defined as the time from sampling to death (uncensored) or last follow up (censored). Dichotomous variables were compared between different groups with the use of the chi-square test. The Mann-Whitney-U-test was used to compare 2 and the Kruskal-Wallis test to compare more than 2 unmatched groups when continuous variables were not normally distributed. Results were considered significant at P < 0.05. Statistical analyses were performed with the SPSS version 19.0.0 (SPSS Inc); the reported P values were 2-sided. Evolution plots were generated according to Miller [12].
3. Results
3.1 Impact of RAS-pathway mutations on the phenotype of CMML
The phenotype stratified by the presence or absence of RASopathy gene mutations in patients with CMML and a TET2/SRSF2 comutation is shown in Table 1. Patients with additional RASopathy gene mutations had a higher WBC count, a lower Hb level, and a lower platelet value. Moreover, splenomegaly and signs of transformation were more frequent in patients with RAS-pathway activation.
3.2 Impact of RAS-pathway mutations on survival of CMML patients
Figure 1 shows the Kaplan-Meier Plots of overall survival in CMML patients stratified by the presence or absence of RASopathy gene mutations in the molecularly defined subgroup of patients with a TET2/SRSF2 comutation (Figure 1A) and patients without such a comutation (Figure 1B). The median survival of CMML patients with mutations in the RAS-pathway components was 15 months as compared to 38 months in patients without such mutations (p=0.0026). Interestingly, the presence of RASopathy gene mutations was not associated with a poor outcome in CMML patients without a TET2/SRSF2 comutation (Figure 1B). In order to determine the relation of the prognostic impact of molecular RAS-pathway aberrations to other established prognostic factors, due to the relatively small sample size, several Cox regression analyses were performed due to the including WBC, Hb, PLT and PB blasts. As shown in Table S2 RASopathy gene mutations retained their significant prognostic impact in the presence of all these parameters.
3.3 Impact of RAS-pathway mutations on spontaneous myeloid colony formation
In vitro cultures data were available from 62 patient samples. We recently were able to show that growth factor independent CFU-GM formation may be a functional surrogate of RAS-pathway activation [13, 14]. The spontaneous formation of CFU-GM in normal individuals (median 4.8/105 PBMNC, range 3.5–8.5) has been reported by us previously [15]. The numbers of spontaneously formed CFU-GM in CMML patients stratified by the presence or absence of RASopathy gene mutations in the molecularly defined subgroup of CMML patients with a TET2/SRSF2 comutation is indicated in Figure 2. The boxplots show a large variation in colony numbers between single patients in the two cohorts, however, median CFU-GM numbers per 105 MNC were significantly higher in TET2/SRSF2-mutant CMML patients with RAS-pathway mutations (md 48, range 0-440, n=30) as compared to patients without additional RAS-pathway aberrations (md 6, range 0-398, n=32; p=0.0048). A CFU-GM number above 100/105 MNC (high colony growth) has been demonstrated by us to be associated with inferior survival in patients with CMML [16]. The proportion of patient samples with high growth factor independent myeloid colony formation was 37% (11/30) and 6% (2/32), respectively (p=0.0033).
3.4 Correlation of RAS-pathway mutations with spontaneous myeloid colony formation
In one CMML patient serial determinations of the mutational landscape during the course of disease and in vitro colony formation were performed. Because of thrombocytopenia and leukopenia the patient had bone marrow puncture in 2006. At that time the patient had a WBC count of 3.1 G/L and 18% monocytes and somatic mutations in TET2 and SRSF2 determined by NGS. Retrospectively, the patient would be classified as oligomonocytic CMML which is defined by persistent (3 months) peripheral blood monocytosis of 0.5-0.9 G/L plus relative monocytosis of >10% of circulating leukocytes [3]. Moreover, NGS showed an additional abnormality in RUNX1 which we could also detect in the hair follicles of the patient suggesting a germline alteration. In 2/2016 the WBC count increased to 16.6 G/L and in 8/2016 to 24.6 G/L. NGS analysis performed at these time points revealed the emergence of a NRAS mutant clone with a VAF of 19% and 41%, respectively. Finally, in vitro cultures, in which spontaneous myeloid colony formation within the normal range was seen in 2/2011, showed highly increased CFU-GM numbers of 170 and 381 /105 MNC, respectively, thereafter. Thus, we can directly demonstrate that the evolution of an NRAS clone was paralleled by the expansion of a growth factor independent clone of myeloid progenitor cells (Figure 3).
|
Parameter |
TET2/SRSF2 without RASopathy mutations N=50 |
TET2/SRSF2 with RASopathy Mutations N=37 |
p-Value |
|
Age |
74.5 (50-92) |
77 (55-93) |
0.1936 |
|
Sex (% males) |
29/50 (58%) |
28/37 (76%) |
0.0864 |
|
WBC G/L |
12.8 (2.9-121.5) |
22.3 (3.8-86.7) |
0.0151 |
|
Hb g/dL |
11.9 (8.1-15.1) |
10.8 (7.0-15.3) |
0.0188 |
|
Plt G/L |
108 (22-448) |
67 (16-309) |
0.0071 |
|
PB Blasts (% present) |
8/45 (17%) |
12/34 (35%) |
0.0763 |
|
Splenomegaly |
4/23 (17%) |
9/15 (60%) |
0.0068 |
|
Transformation |
2/50 (4%) |
7/37 (19%) |
0.0239 |
Table 1: The phenotype stratified by the presence or absence of RASopathy gene mutations in patients with CMML and a TET2/SRSF2 comutation.
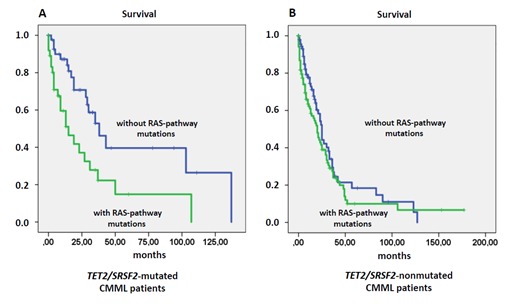
Figure 1: Overall survival in TET2/SRSF2-comutated (Figure 1A) and in TET2/SRSF2-nonmutated CMML patients stratified by the presence or absence of RAS-pathway mutations.
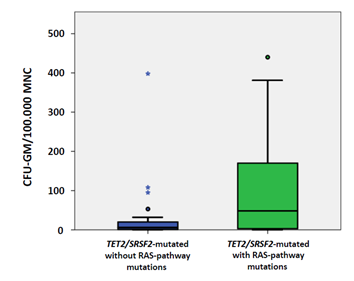
Figure 2: Box plots showing the distribution of spontaneous myeloid colony numbers in TET2/SRSF2-mutant CMML patients with or without additional mutations in RASopathy genes including median values, minimum values, maximum values, as well as upper and lower quartiles, respectively. Cultures were plated in duplicates or triplicates, respectively, at 25-100 × 103 PBMNC/mL. Aggregates with more than 40 translucent, dispersed cells were counted as CFU-GM. CFU-GM data from patients are expressed as mean values from cultures.
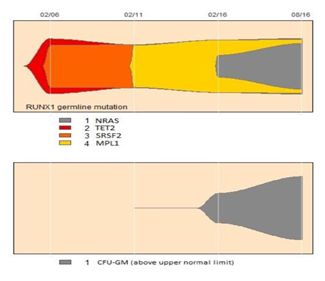
Figure 3: Evolution plot of a CMML patient with TET2/SRSF2 comutation and NRAS mutation who had also serial determinations of spontaneous in vitro myeloid colony formation.
4.Discussion
The RAS signaling pathway is one of the major signaling pathways which are involved into the the transduction of growth stimulatory signals from GF receptors to the nucleus where cell programs associated with cell growth are activated. The “RASopathies” are a group of genetic syndromes caused by germline mutations in genes that encode components of the RAS signaling pathway including NRAS, KRAS, NF-1, CBL, and PTPN11 [17-22]. Besides their developmental defects they share a predisposition to juvenile myelomonocytic leukemia, a hematologic malignancy of early childhood. Moreover, spontaneous molecular aberrations of components of this pathway are the most frequently mutated genes in cancers. In the preclinical mouse model myelomonocytic leukemias can be recapitulated by transplantation of mouse BM cells harboring an oncogenic mutation in the Nras locus [23]. Interestingly, alterations of the other RASopathy genes may also lead to a similar phenotype in preclinical mouse models [24-28]. Mice develop a myeloproliferative disorder with clonal expansion of the granulomonopoiesis in vivo and show spontaneous in vitro myeloid colony formation without exogenous growth factors due to aberrant GM-CSF signaling.
Despite a number of studies investigating the role of the RAS signaling pathway in CMML remains controversial. There are studies showing that mutations RAS components are associated with inferior survival but there are also studies which failed to show this. The presence or absence of a RAS point mutation was not a significant prognostic parameter in a univariate analysis from 66 CMML patients reported by Onida in 2002 [5]. In univariate analysis OS rates decreased in 312 molecularly characterized CMML patients with mutations in CBL and there was a trend towards shorter survival in NRAS mutated patients in a study by Itzykson [6]. Since NRAS mutations significantly affected survival in univariate analysis and retained its prognostic value in multivariable Cox regression mutations in NRAS were integrated in the risk assessment of 214 patients with CMML by Elena [7]. An international dataset analysed by Padron revealed in addition to ASXL1 mutations mutations in CBL but not in NRAS or KRAS as a new molecular independent prognostic parameter [8]. Restricting analysis to molecularly defined subgroups may be more appropriate to study the effects of RAS pathway mutations on the phenotype and clinical outcome. In the preclinical mouse model it is possible to work with clearly defined molecular categories. Im mice with a TET2-mutant background, the transfection of NRAS mutation convincingly aggravated the disease [29, 30]. Here we show in CMML cells from patients with TET2/SRSF2 comutation that the presence of mutations in RASopathy genes clearly changed the phenotype of disease and shortened survival.
The basis for all RAS pathway-oriented treatment concepts is the identification of RAS pathway hyperactivation in patients. Due to the fact that in CMML more than one molecular aberration can be detected in the majority of patients, functional tests may be important to better estimate the contribution of a particular molecular aberration in the pathogenesis of the malignancy. We recently were able to show that spontaneous CFU-GM formation may be a functional surrogate of RAS pathway activation [12, 13]. Hyperactivation of the RAS signaling pathway in this study was not only demonstrated at the molecular level but also at a functional level by comparing the number of growth factor independent myeloid colony formation in CMML patients with the TET2/SRSF2 comutation and the presence or absence of additional mutations in RASopathy genes. Spontaneous colony formation was much higher in patients with molecular aberrations of the RAS signaling pathway as compared to patients without these aberrations. In one patient who had serial determinations of the mutational landscape and in vitro colony formation we can directly demonstrate that the evolution of an NRAS clone was paralleled by increasing myeloid colony growth.
The clinical implication of our findings is supported by the recent development of novel RAS pathway inhibitors [31]. Since RAS is the most frequently mutated gene family in cancers, investigators have sought an effective RAS inhibitor for more than three decades. RAS inhibitors, however, were so elusive that RAS was termed ‘undruggable’. With the recent success of allele- specific covalent inhibitors we have now the opportunity to evaluate the best therapeutic strategies to treat RAS-driven cancers. Inhibition of the RAS pathway with or without other agents etsablished in the treatment of myeloid malignancies may be attractive approaches to treat RAS- mutant CMML.
Authors Contributions
K.G. directed the research, collected, analyzed and interpreted the data and wrote the manuscript; E.J. performed colony assays; A.B., M.G. performed NGS analyses; T.G. and E.G. performed the administration of data; G.H. interpreted molecular data; T.N., M.P., S.M.,R.S., A.Z,. H.S., L.Ö., R.K., P.V, provided patient samples and clinical information; All authors had the opportunity to review the manuscript.
Funding
This study was supported by the “Gesellschaft zur Erforschung der Biologie und Therapie von Tumorkrankheiten” – ABCMML-112015” and the “Austrian Science Fund (FWF) – grant F4704-B20”.
Conflict of Interest Disclosure
The authors declare no conflict of interest.
References
- Coltro G, Patnaik MM. Chronic Myelomonocytic Leukemia: Insights into Biology, Prognostic Factors, and Treatment. Curr Oncol Rep 21 (2019): 101.
- Itzykson R, Fenaux P, Bowen D, et al. Diagnosis and Treatment of Chronic Myelomonocytic Leukemias in Adults: Recommendations From the European Hematology Association and the European LeukemiaNet. HemaSphere 2 (2018): e150.
- Valent P, Orazi A, Savona MR, et al. Proposed diagnostic criteria for classical chronic myelomonocytic leukemia (CMML), CMML variants and pre-CMML conditions. Haematologica 104 (2019): 1935-1949.
- Patnaik MM, Tefferi A. Cytogenetic and molecular abnormalities in chronic myelomonocytic leukemia. Blood Cancer Journal 6 (2016): e393.
- Onida F, Kantarjian HM, Smith TL, et al. Prognostic factors and scoring systems in chronic myelomonocytic leukemia: a retrospective analysis of 213 patients. Blood 99 (2002): 840-849.
- Itzykson R, Kosmider O, Renneville A, et al. Prognostic Score Including Gene Mutations in Chronic Myelomonocytic Leukemia. Journal of Clinical Oncology 31 (2013): 2428-2436.
- Elena C, Gallì A, Such E, et al. Integrating clinical features and genetic lesions in the risk assessment of patients with chronic myelomonocytic leukemia. Blood 128 (2016): 1408-1417.
- Padron E, Garcia-Manero G, Patnaik MM, et al. An international data set for CMML validates prognostic scoring systems and demonstrates a need for novel prognostication strategies. Blood Cancer Journal 5 (2015): e333.
- Geissler K, Jäger E, Barna A, et al. The Austrian biodatabase for chronic myelomonocytic leukemia (ABCMML): A representative and useful real-life data source for further biomedical research. Wien Klin Wochenschr 131 (2019): 410-418.
- Ohler L, Geissler K, Hinterberger W. Diagnostic and prognostic value of colony formation of hematopoietic progenitor cells in myeloid malignancies. Wien Klin Wochenschr 115 (2003): 537-546.
- Geissler K, Ohler L, Födinger M, et al. Interleukin 10 inhibits growth and granulocyte/macrophage colony-stimulating factor production in chronic myelomonocytic leukemia cells. J Exp Med 184 (1996): 1377-1384.
- Miller CA, McMichael J, Dang HX, et al. Visualizing tumor evolution with the fishplot package for R. BMC Genomics 17 (2016): 880.
- Geissler K, Jäger E, Barna A, et al. Chronic myelomonocytic leukemia patients with RAS pathway mutations show high in vitro myeloid colony formation in the absence of exogenous growth factors. Leukemia 30 (2016): 2280-2281.
- Geissler K, Jäger E, Barna A, et al. Correlation of RAS-Pathway Mutations and Spontaneous Myeloid Colony Growth with Progression and Transformation in Chronic Myelomonocytic Leukemia-A Retrospective Analysis in 337 Patients. IJMS 21 (2020): 3025.
- Oehler L, Foedinger M, Koeller M, et al. Interleukin-10 inhibits spontaneous colony-forming unit-granulocyte-macrophage growth from human peripheral blood mononuclear cells by suppression of endogenous granulocyte-macrophage colony-stimulating factor release. Blood 89 (1997): 1147-1153.
- Sagaster V, Ohler L, Berer A, et al. High spontaneous colony growth in chronic myelomonocytic leukemia correlates with increased disease activity and is a novel prognostic factor for predicting short survival. Ann Hematol 83 (200): 9-13.
- Shannon KM, O'Connell P, Martin GA, et al. Loss of the normal NF1 allele from the bone marrow of children with type 1 neurofibromatosis and malignant myeloid disorders. N Engl J Med 330 (1994): 597-601.
- Kalra R, Paderanga DC, Olson K, et al. Genetic analysis is consistent with the hypothesis that NF1 limits myeloid cell growth through p21ras. Blood 84 (1994): 3435-3439.
- Tartaglia M, Niemeyer CM, Fragale A, et al. Somatic mutations in PTPN11 in juvenile myelomonocytic leukemia, myelodysplastic syndromes and acute myeloid leukemia. Nat Genet 34 (2003): 148-150.
- Loh ML, Vattikuti S, Schubbert S, et al. Mutations in PTPN11 implicate the SHP-2 phosphatase in leukemogenesis. Blood 103 (2004): 2325-2331.
- Niemeyer CM, Kang MW, Shin DH, et al. Germline CBL mutations cause developmental abnormalities and predispose to juvenile myelomonocytic leukemia. Nat Genet 42 (2010): 794-800.
- Niemeyer CM. RAS diseases in children. Haematologica 99 (2014): 1653-1662.
- Wang J, Liu Y, Li Z, et al. Endogenous oncogenic Nras mutation promotes aberrant GM-CSF signaling in granulocytic/monocytic precursors in a murine model of chronic myelomonocytic leukemia. Blood 116 (2010): 5991-6002.
- Van Meter ME, Díaz-Flores E, Archard JA, et al. K-RasG12D expression induces hyperproliferation and aberrant signaling in primary hematopoietic stem/progenitor cells. Blood 109 (2007): 3945-3952.
- Li Q, Haigis KM, McDaniel A, et al. Hematopoiesis and leukemogenesis in mice expressing oncogenic NrasG12D from the endogenous locus. Blood 117 (2011): 2022-2032.
- Parikh C, Subrahmanyam R, Ren R. Oncogenic NRAS rapidly and efficiently induces CMML-and AML-like diseases in mice. Blood 108 (2006): 2349-2357.
- Chan RJ, Leedy MB, Munugalavadla V, et al. Human somatic PTPN11 mutations induce hematopoietic-cell hypersensitivity to granulocyte-macrophage colony-stimulating factor. Blood 105 (2005): 3737-3742.
- Le DT, Kong N, Zhu Y, et al. Somatic inactivation of Nf1 in hematopoietic cells results in a progressive myeloproliferative disorder. Blood 103 (2004): 4243-4250.
- Kunimoto H, Meydan C, Nazir A, et al. Cooperative Epigenetic Remodeling by TET2 Loss and NRAS Mutation Drives Myeloid Transformation and MEK Inhibitor Sensitivity. Cancer Cell 33 (2018): 44-59.
- Jin X, Qin T, Zhao M, et al. Oncogenic N-Ras and Tet2 haploinsufficiency collaborate to dysregulate hematopoietic stem and progenitor cells. Blood Adv 2 (2018): 1259-1271.
- Moore AR, Rosenberg SC, McCormick F, et al. RAS-targeted therapies: is the undruggable drugged? Nat Rev Drug Discov 19 (2020): 533-552.


 Impact Factor: * 4.1
Impact Factor: * 4.1 Acceptance Rate: 74.74%
Acceptance Rate: 74.74%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 