WISP1 and Mir-29c-3p are Novel Prognostic Biomarkers and Therapeutic Targets for Bladder Cancer
Zhou Hongyi1, Zhu Ming1, Zhu Leilei1, Xu Zhuoqun1, Wang Zhuo2*, Shao Jianfeng1*
1Department of urology, The Affiliated Wuxi People's Hospital of Nanjing Medical University, Wuxi 214023, Jiangsu Province, China
2Department of Geriatrics, The Affiliated Wuxi People's Hospital of Nanjing Medical University, Wuxi 214023, Jiangsu Province, China
*Corresponding Authors: Shao Jianfeng, Professor, Department of Geriatrics, The Affiliated Wuxi People's Hospital of Nanjing Medical University, Wuxi 214023, Jiangsu Province, China
Wang Zhuo, Department of Geriatrics, The Affiliated Wuxi People's Hospital of Nanjing Medical University, Wuxi 214023, Jiangsu Province, China
Received: 10 November 2021; Accepted: 22 November 2021; Published: 07 December 2021
Article Information
Citation:
Hongyi Z, Ming Z, Leilei Z Zhuoqun X, Zhuo W, Jianfeng S. WISP1 and Mir-29c-3p are Novel Prognostic Biomarkers and Therapeutic Targets for Bladder Cancer. Journal of Cancer Science and Clinical Therapeutics 5 (2021): 520- 531.
View / Download Pdf Share at FacebookAbstract
Aberrant expression of WISP1 is associated with carcinogenesis; however, the expression and prognostic values of WISP1 in bladder cancer remain elusive. Therefore, the present study aimed to investigate the WISP1 expression in bladder cancer and explore the possible mechanisms and clinical value of WISP1 in bladder carcinogenesis. The in silico analysis based on oncomine and Kaplan-Meier Plotter databases revealed that WISP1 was up-regulated in bladder cancer and associated with poor survival in patients with bladder cancer. Besides, WISP1 expression was identified to be up-regulated both in bladder cancer tissues and cell lines by qRT-PCR and Western blot assays. Using TargetScan database, we identified 13 miRNAs that putatively regulate the expression of WISP1. Of these miRNAs, only miR-29c-3p was found to be significantly negatively correlated with WISP1 in bladder cancer tissues. The correlation of in situ expressions of WISP1 and miR-29c-3p by immunohistochemistry (IHC) and clinical characteristics revealed that WISP1 was significantly associated with tumor size and hsa-miR-23b- 3p expression, and miR-29c-3p was associated with tumor size, M stage, and WISP1 expression. Multivariate Cox regression analysis indicated that TNM stage and WISP1 expression were predictors of unfavorable prognosis, while hsa-miR-29c-3p was a predictor of favorable prognosis in patients with bladder cancer. Collectively, the findings indicated that WISP1 and miR-29c-3p might serve as novel prognostic biomarkers and potential therapeutic targets for bladder cancer.
Keywords
<p>BLAD; Bladder cancer; miR-29c-3p; Prognostic biomarkers; WISP1</p>
Article Details
1. Introduction
Globally, (BC) ranks the tenth most frequently diagnosed urological malignancy, with approximately 550,000 new cases (nearly 425,000 in males and 125,000 in females) diagnosed in 2018 worldwide. It is among the leading causes of cancer-associated mortality, with approximately 200,000 deaths reported in 2018. Histologically, urothelial carcinoma (transitional cell) represents the predominant type of bladder cancer, accounting for 90 percent of all bladder cancers [1]. Clinically, two main phenotypes, including the muscle-invasive and non-muscle-invasive, with different pathogenesis, molecular characteristics, and clinical outcomes, have been identified [2,3]. Recent advances in high-throughput sequencing technologies have facilitated the rapid molecular characterization and enhanced our understanding of the pathogenesis of bladder cancer, leading to the identification of actionable therapeutic targets for bladder cancer [4]. However, the molecular mechanisms underlying the occurrence and development of bladder carcinogenesis remain to be completely elucidated.
WNT1-Inducible Signaling Pathway Protein 1 (WISP1), a secreted matricellular protein, is a member of the connective tissue growth factor/CCN protein family, which is found in the Extracellular Matrix (ECM). WISP1 is predominantly involved in various biological processes, including cell adhesion, proliferation, differentiation, survival, and carcinogenesis. The elevated expression of WISP1 has been detected in several cancers, including melanoma, glioblastoma, and hepatocellular carcinoma [5-7]. WISP1 expression has also been associated with tumor purity, an inflamed tumor microenvironment, advanced disease, EMT, and macrophage M2 polarization in multiple solid tumors [8-11].
However, its expression and physiological function in bladder cancer remain elusive. MicroRNAs (miRNAs) are small 18–24 nucleotide long, single-stranded non-coding RNAs involved in post-transcriptional gene regulation by pairing to the mRNAs of its target protein-coding gene), thereby causing translational repression or mRNA degradation of the target gene [12]. Recently, several miRNAs, such as miRNA-217 and miRNA-616 [13,14], have been identified as prognostic biomarkers in bladder cancer [15,16]. However, there is a paucity of studies on WISP1-related miRNAs in bladder cancer. Therefore, in the present study, we investigated the expression levels of WISP1 in bladder cancer using quantitative real-time PCR (qRT-PCR), Western blot assay, and bioinformatics analyses. Furthermore, we used TargetScan to predict the potential WISP1-related miRNAs. Univariate and multivariate COX regression and the Kaplan-Meier method were applied to evaluate the prognostic value of WISP1 and miRNAs in bladder cancer.
2. Materials and Methods
2.1 Bioinformatics analysis
Oncomine (www.oncomine.org), an online microarray database, was used to analyze the mRNA levels of WISP1 in bladder cancer and normal bladder tissue samples [17,18]. Kaplan-Meier Plotter (kmplot.com/analysis/) was used to analyze the prognostic value of WISP1 in bladder cancer [19]. TargetScan (7.2 version, http://www.targetscan.org/) was used to identify the potential miRNAs targetingWISP1 [20].
2.2 BLAD Patient Samples
One hundred and thirty-two BC tissues and twenty paired bladder cancer tissues and adjacent normal tissues were obtained from patients who underwent surgical treatment for either diagnostic biopsy or surgical resection for bladder cancer at Wuxi People's Hospital Affiliated to Nanjing Medical University. The study protocols were approved by the Ethics Committee of the Institutional Review Board of Wuxi People's Hospital, Nanjing Medical University. Written informed consent was obtained from each patient. All protocols involving human patients were performed in accordance with the relevant guidelines and regulations of Wuxi People's Hospital, Nanjing Medical University. The tissue samples were immediately stored at −80°C until use. The histopathological examination was performed to confirm the diagnosis.
2.3 Cell lines and cell culture
The human bladder cancer cell lines, including SV-HUC-1, UMUC3, RT4, T24, UC9, and 5637, were obtained by American Type Culture Collection (ATCC; Manassas, VA, USA). Cells were cultured in the Dulbecco's Modifed Eagle's Medium (Life Technologies) supplemented with 10% fetal bovine serum (FBS; Invitrogen), 10% (100µg/mL) penicillin, and 100 U/mL streptomycin in a humidified atmosphere 5% CO2 at 37°C.
2.4 Quantitative reverse transcription PCR (RT-qPCR)
Total RNA was extracted from fresh frozen tissue samples using Trizol reagent (Invitrogen, Carlsbad, CA, United States) according to the manufacturer's instructions. cDNA was synthesized using a reverse transcription kit (PrimeScript RT-PCR kit; Takara, Japan) following the manufacturer's protocol.Total RNA was isolated from the cells using the PureLink™ RNA Mini Kit (Life Technologies, Carlsbad, CA, USA) according to the manufacturer′s instructions. Reverse transcription reactions were performed using iScript cDNA synthesis kit (Bio-Rad) for mRNA, or TaqMan miR Reverse Transcription kit (Applied Biosystems) for mature miR as described earlier. qRT-PCR was performed using SYBR Green Master Mix (Takara, Japan) on an ABI 7500 RealTime PCR System (Applied Biosystems, United States). β-actin and U6 were used as the reference genes. Primers used in the study were listed in Table1. All reactions were performed in triplicate. The relative expression levels of miRNA and mRNA were evaluated using the 2-??Ct method.
|
Names |
Forward |
Reverse |
|
WISP1 |
5′-GAAGCAGTCAGCCCTTATG-3′ |
5′-CTTGGGTGTAGTCCAGAAC-3′ |
|
β-actin |
5′-CCTGTGGCATCCACGAAACT-3′ |
5′-GAAGCATTTGCG GTGGACGAT-3′ |
|
U6 |
5′-CTCGCTTCGGCAGCACA-3′ |
5′-AACGCTTCACGAATTTGCGT-3′ |
Table 1: Primers of genes and miRNAs.
2.5 Western blot assay
Total cell proteins were extracted from bladder cancer, and paracancerous tissues with RIPA lysis buffer (Pierce, Thermo Scientific, Cramlington, United Kingdom) supplemented with protease inhibitor cocktail. Protein concentrations were determined by BCA protein concentration reagent kit (Beyotime, China). Proteins were separated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDA-PAGE) and transferred to PVDF membrane (Bio-Rad, USA). The membranes were blocked with 5% non-fat dry milk for 1 h at room temperature (RT). Subsequently, the membranes were incubated with the following primary antibodies: anti-WISP1 (Abcam, ab178547) and anti-β-actin (Abcam, ab8226) at 4°C for overnight. Then, membranes were washed and incubated with goat anti-rabbit IgG secondary antibody conjugated with horseradish peroxidase (1:1000). The target bands were visualized using an electrochemiluminescence (ECL) detection system. β-actin was used as an endogenous control.
2.6 Immunohistochemistry (IHC)
The immunohistochemical assay was performed on cancer tissues and adjacent normal tissues from bladder cancer patients. Tissue specimens were fixed with 4% paraformaldehyde, dehydrated, and embedded in paraffin. The tissue section was cut into a thickness of 5 μm and then mounted on a glass slide. IHC staining was performed according to the manufacturer's protocol. In brief, slides were deparaffinized and then rehydrated in successively graded ethanol. Antigen retrieval was performed by heating the sections at 100°C for 30 minutes in the microwave. Subsequently, the slides were incubated with the primary antibody against WISP1 (diluted 1:200, ab178547, Abcam, USA) at 4°C overnight. The sections were washed 3 times in TBST for 5 minutes each and then incubated with the rabbit secondary antibody for 1 hour at room temperature. The sections were then stained with diaminobenzidine (DAB) and counterstained with hematoxylin. Three pathologists independently performed blinded analysis of the WISP1 immunostaining intensity under a light microscope. By analyzing the Based on the percentage of positively staining cells, the sections were grade as 0 = 5% or none of the cells were stained, 1 = 6-25% of the cells stained positive, 2 = 26-50% of the cells stained positive; 3 = 51-75% of the cells stained positive, and 4 = more than 76% of the cells stained positive. Similarly, the staining intensity was graded as 0 = no staining, 1 = weak staining, 2 = moderate staining, and 3 = strong staining. The two scores were multiplied, and the immune-reactive score (values from 0-12) for each case was determined [21].
3. Statistical Analysis
All experiments were performed in triplicate. Data from three or more independent experiments were presented as the mean ± standard deviation (SD). A paired Student's t-test was used to analyze the final score of cancer tissues and adjacent normal tissues. A Chi-square test was performed to assess the relationship between WISP1 and miRNAs expression and clinicopathological characteristics. The Kaplan-Meier method was used to calculate survival functions, and differences were compared using the log-rank test. Univariate and multivariate Cox proportional hazards analyses were used to identify the independent prognostic factors for overall survival in bladder cancer. Statistical analyses were performed with the R version 3.6.1 (R Core Team, 2019) with RStudio and GraphPad Prism 8.0 (GraphPad Software, San Diego, CA, United States). A P-value of < 0.05 (two-tailed) was considered statistically significant.
4. Results
4.1 WISP1 was over expressed in bladder cancer
From the oncomine database, as illustrated in Figure 1a, we determined that WISP1 was up-regulated in bladder cancer. From the Kaplan-Meier plotter database, as presented in Figure 1b, the high expression of WISP1 was significantly associated with poor overall survival. These results indicated that the aberrant expression of WISP1 in bladder cancer was associated with unfavorable survival. qRT-PCR and Western blot analyses of cancer tissues and adjacent normal tissues from bladder cancer patients revealed that WISP1 was noticeably up-regulated in bladder cancer tissues, as shown in Figure 1c, d/e. Consistently, a significant over-expression of WISP1 was observed in bladder cancer cells (UMUC3, RT4, T24, UC9, and 5637) compared to normal cell lines (SV-HUC-1) as represented in Figure 1f/g. Collectively, these results suggested that WISP1 was significantly up-regulated in bladder cancer.
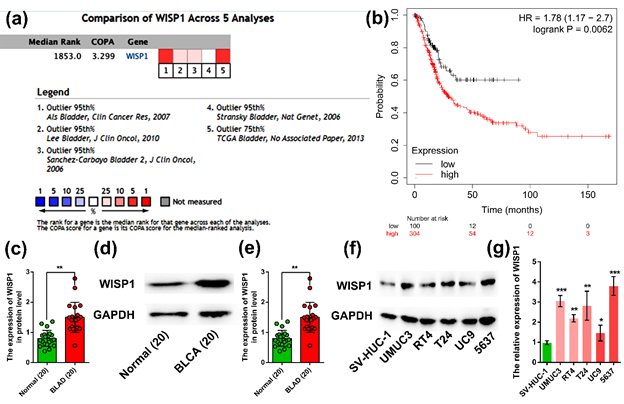
(a) The expression of WISP1 in bladder cancer using oncomine database; (b) Association of WISP1 expression with overall survival in bladder cancer using Kaplan-Meier plotter database; (c) WISP1 mRNA expression in bladder cancer tissues and adjacent normal tissues by qRT-PCR; (d) Western blot analysis of bladder cancer tissues and adjacent normal tissues; (e) Results of the protein expression of WISP1 in bladder cancer tissues and adjacent normal tissues by Western blot assay; (f) Results of the protein expression of WISP1 in normal and bladder cancer cell lines by Western blot assay; (g) Statistical analyses of the protein expression of WISP1 in normal and bladder cancer cell lines by Western blot assay. ***represented P < 0.001; **represented P < 0.01; *represented P < 0.05.
Figure 1: WISP1 overexpression in bladder cancer.
4.2 hsa-miR-29c-3p expression was negatively associated with WISP1 expression
To explore potential miRNAs related to WISP1, we used the TargetScan and identified 13 miRNAs according to their conserved sites that putatively regulate the expression of WISP1 (Figure 2a). Among these miRNAs, hsa-miR-23a-3p and hsa-miR-29b-3p were markedly up-regulated, while hsa-miR-16-5p, hsa-miR-23b-3p, and hsa-miR-29c-3p were significantly down-regulated in bladder cancer compared to its normal adjacent tissues (Figure 2b). However, only hsa-miR-23b-3p was significantly negatively correlated with WISP1, while the others were not (Figure 2c/d/f). These results indicated that hsa-miR-29c-3p was significantly negatively correlated with WISP1 expression in bladder cancer.
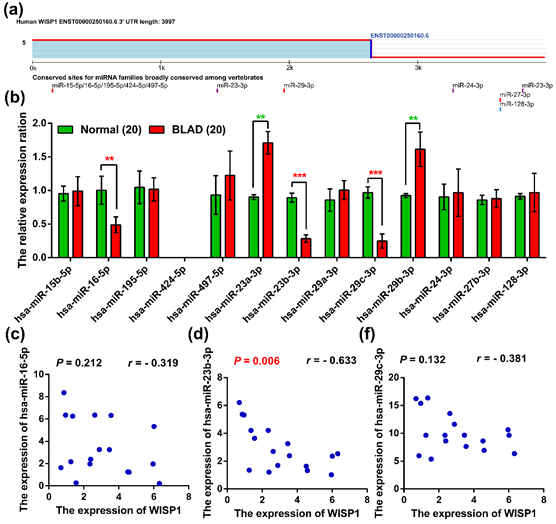
(a)Conserved target sites of miRNA families for WISP1 using TargetScan analysis; (b) The expression of related miRNAs in normal and bladder cancer tissues; (c, d, f) The related expression of WISP1. ***represented P < 0.001; **represented P < 0.01.
Figure 2: The miRNAs expression was associated with WISP1 in bladder cancer.
4.3 WISP1 was associated with tumor size and hsa-miR-23b-3p expression
To further verify WISP1 expression in clinical specimens of bladder cancer, IHC and qRT-PCR was performed in 132 bladder cancer tissue specimens. The protein expression of WISP1 was localized both in the nucleus and cytoplasm of cells (Figure3b). The low expression of WISP1 was detected in 92 samples and 40 samples exhibited high expression of WISP1 (figure3a/b). The expression level of hsa-miR-23b-3p in the high expression group of WISP1 was evidently lower than that in the low group by Student's t-test (Figure3c). The correlation analysis of WISP1 and hsa-miR-23b-3p expression with clinical characteristics revealed that tumor size and hsa-miR-23b-3p expression were significantly associated with WISP1 expression; however, age, gender, T stage, N stage, M stage, TNM stage, and differentiation were not significantly associated with WISP1 expression (Table 2). Moreover, tumor size, M stage, and WISP1 were significantly correlated with hsa-miR-23b-3p expression; however, age, gender, T Stage, N Stage, TNM stage and differentiation were not associated with hsa-miR-23b-3p expression (Table2). These findings indicated that WISP1 and hsa-miR-23b-3p might play an important role in bladder cancer.
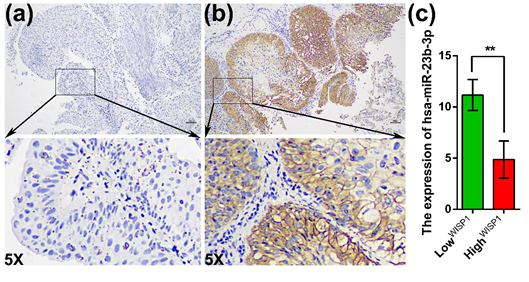
(a) The low expression of WISP1; (b) The high expression of WISP1; (c) The expression of hsa-miR-23b-3p in the groups of low and high expression of WISP1. 5X indicates magnification of 5X.
Figure 3: The correlation between WISP1 and hsa-miR-23b-3p expression.
|
Characteristics |
No. |
WISP1 |
hsa-miR-23b-3p |
||||
|
Low |
High |
P value |
Low |
High |
P value |
||
|
Age |
0.27 |
0.128 |
|||||
|
<=60 |
78 |
51 |
27 |
33 |
45 |
||
|
>60 |
54 |
41 |
13 |
15 |
39 |
||
|
Gender |
0.119 |
0.881 |
|||||
|
Female |
74 |
47 |
27 |
26 |
48 |
||
|
Male |
58 |
45 |
13 |
22 |
36 |
||
|
T Stage |
0.239 |
0.42 |
|||||
|
T1-T2 |
68 |
51 |
17 |
22 |
46 |
||
|
T3-T4 |
64 |
41 |
23 |
26 |
38 |
||
|
N Stage |
0.699 |
0.229 |
|||||
|
N0 |
61 |
41 |
20 |
26 |
35 |
||
|
N1-N3 |
71 |
51 |
20 |
22 |
49 |
||
|
M Stage |
0.141 |
0.006 |
|||||
|
M0 |
55 |
34 |
21 |
28 |
27 |
||
|
M1 |
77 |
58 |
19 |
20 |
57 |
||
|
TNM Stage |
0.21 |
0.14 |
|||||
|
I-II |
45 |
35 |
10 |
12 |
33 |
||
|
III-IV |
87 |
57 |
30 |
36 |
51 |
||
|
Differentiation |
0.204 |
0.054 |
|||||
|
Poor and Middle |
66 |
42 |
24 |
30 |
36 |
||
|
Well |
65 |
49 |
16 |
18 |
47 |
||
|
Tumor Size |
0.011 |
0.021 |
|||||
|
<=4 cm |
60 |
49 |
11 |
15 |
45 |
||
|
>4 cm |
72 |
43 |
29 |
33 |
39 |
||
|
hsa-miR-29c-3p |
<0.001 |
||||||
|
Low |
48 |
17 |
31 |
||||
|
High |
84 |
75 |
9 |
||||
|
WISP1 |
<0.001 |
||||||
|
Low |
92 |
17 |
75 |
||||
|
High |
40 |
31 |
9 |
Table 2: The correlation between WISP1 expression and clinicopathological characteristics in bladder cancer.
4.4 WISP1 and hsa-miR-23b-3p were independent prognostic factors for bladder cancer
Using Kaplan-Meier survival analysis, BLAD patients with high WISP1 expression exhibited a significantly low OS (P = 0.011, Figure 4a); BLAD patients with high hsa-miR-23b-3p expression exhibited a noticeably good OS (P = 0.015, Figure 4b). The results of the univariate analysis indicated that age (>60 vs. <=60), M stage (M1 vs. M0), TNM stage (III-IV vs. I-II), hsa-miR-29c-3p (High vs. Low), and WISP1 (High vs. Low) were critical factors affecting the overall survival in patients with bladder cancer (P < 0.05, Table 2). The results of multivariate Cox regression analysis showed that TNM stage (III-IV vs. I-II) and WISP1 (High vs. Low) were independent predictors of unfavorable prognosis, while hsa-miR-29c-3p (High vs. Low) represented a predictor of favorable prognosis in patients with bladder cancer (P < 0.05, Table 3). Together, these results revealed that WISP1 and hsa-miR-23b-3p were independent prognostic factors of bladder cancer.
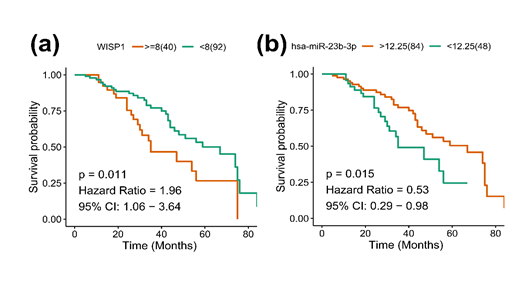
(a) Kaplan-Meier survival curves for WISP1 expression; (b) Kaplan-Meier survival curves for hsa-miR-23b-3p expression.
Figure 4: WISP1 and hsa-miR-23b-3p associated with overall survival of bladder cancer.
|
Characteristics |
Univariate Cox regression analysis |
Multivariate Cox regression analysis |
||||
|
HR |
95% CI |
P Value |
HR |
95% CI |
P Value |
|
|
Age (>60 vs. <=60) |
2.3 |
1.27-4.17 |
0.006 |
1.58 |
0.84-2.97 |
0.155 |
|
Gender(Female vs. Male) |
1.37 |
0.8-2.35 |
0.252 |
|||
|
Tumor Size (>4 cm vs. <=4 cm) |
0.84 |
0.49-1.44 |
0.518 |
|||
|
Differation (Poorly vs. Well/moderately) |
0.91 |
0.53-1.56 |
0.733 |
|||
|
T Stage(T3-T4 vs. T1-T2) |
1.25 |
0.73-2.14 |
0.413 |
|||
|
N Stage(N1-N3 vs. N0) |
1.05 |
0.61-1.8 |
0.862 |
|||
|
M Stage(M1 vs. M0) |
1.94 |
1.13-3.31 |
0.016 |
1.61 |
0.94-2.76 |
0.085 |
|
TNM Stage(III-IV vs. I-II) |
2.58 |
1.48-4.48 |
0.001 |
2.71 |
1.51-4.89 |
0.001 |
|
hsa-miR-29c-3p (High vs. Low) |
0.29 |
0.16-0.52 |
<0.001 |
0.46 |
0.22-0.96 |
0.038 |
|
WISP1 (High vs. Low) |
2.71 |
1.55-4.73 |
<0.001 |
2.15 |
1.07-4.32 |
0.031 |
Table 3: Univariate and multivariate COX regression analyses of the prognostic factors in bladder cancer.
5. Discussion
WISP1, a cysteine-rich protein, belongs to the family of matricellular proteins involved in developmental functions and carcinogenesis. Furthermore, WISP1 polymorphisms have been recognized as a biomarker or a therapeutic target in urothelial cell carcinoma [22]. From the oncomine database and Kaplan-Meier plotter database, we found that WISP1 was up-regulated in bladder cancer and associated with significantly poor overall survival, suggesting a potential role of WISP1 in bladder cancer. The expression of WISP1, both at mRNA and protein levels, was evaluated in bladder cancer cell lines and tissues, and the results indicated that WISP1 was indeed overexpressed in bladder cancer. miRNAs, critical regulators of gene expression, are often dysregulated in cancer [23-26]. Using TargetScan, we predicted 13 miRNAs, which potentially target the WISP1. However, correlation analysis revealed that only hsa-miR-23b-3p was significantly negatively correlated with WISP1 (P=0.006, r=-0.633) in bladder cancer. Notably, miR-23b-3p has also been reported as a robust normalizer for urine microRNA studies in bladder cancer [27]. Accumulating studies have indicated that miR-23b-3p acts as a tumor suppressor in multiple cancer, including laryngeal squamous cell carcinoma [28], ovarian cancer [29], esophageal Carcinoma [30], and bladder cancer [31]; however, in abeta-treated neuroblastoma cells, miR-23b-3p functions as an oncogenic factor [32].
Furthermore, tumor size and hsa-miR-23b-3p expression were significantly associated with WISP1, and tumor size, M stage and WISP1 were significantly correlated wth hsa-miR-23b-3p expression. All these findings indicated that WISP1 and hsa-miR-23b-3p might play crucial roles in bladder cancer. The results of univariate Cox analysis revealed that age, M stage, TNM stage, hsa-miR-29c-3p, and WISP1 were critical factors affecting the survival time in patients with bladder cancer. The multivariate Cox survival analysis revealed that TNM stage and WISP1were predictors of unfavorable prognosis, while hsa-miR-29c-3p represented a factor of favorable prognosis in patients with bladder cancer. Furthermore, bladder cancer patients with high WISP1 expression exhibited a significantly lower OS, while patients with high hsa-miR-23b-3p expression exhibited an evidently higher OS. Taken together, these results suggested that WISP1 and hsa-miR-23b-3p were independent prognostic factors of bladder cancer.
Several potential limitations of the present study should be noted. Firstly, although validated in databases, our bioinformatics analyses with the current databases may have certain limitations and flaws. However, the evidence supporting the involvement of WISP1 and hsa-miR-23b-3p in bladder cancer remains very limited. Thus, further studies are warranted to validate with mechanistic evidence the designation of WISP1 as a potential target of hsa-miR-23b-3p for bladder cancer. Secondly, the functional mechanism underlying WISP1 overexpression in bladder cancer was not investigated; therefore, further studies are required to identify the mechanisms of WISP1 overexpression in bladder carcinogenesis. In conclusion, the results of the present study indicated that WISP1 and miR-29c-3p may serve as novel prognostic biomarkers and potential therapeutic targets for bladder cancer.
Funding
This work was financially supported by Top Talent Support Program for young and middle-aged people of Wuxi Health Committee (HB2020013)
References
- Grayson M. Bladder cancer. Nature 551 (2017): S33.
- Hurst C, Rosenberg J, Knowles M. SnapShot: Bladder Cancer. Cancer Cell 34 (2018): 350-351.
- Sacher AG, St Paul M, Paige CJ, Ohashi PS. Cytotoxic CD4(+) T Cells in Bladder Cancer-A New License to Kill. Cancer Cell 38 (2020): 28-30.
- Oh DY, Kwek SS, Raju SS, et al. Intratumoral CD4(+) T Cells Mediate Anti-tumor Cytotoxicity in Human Bladder Cancer. Cell 181 (2020): 1612-1625.
- Deng W, Fernandez A, McLaughlin SL, et al. WNT1-inducible signaling pathway protein 1 (WISP1/CCN4) stimulates melanoma invasion and metastasis by promoting the epithelial-mesenchymal transition. J Biol Chem 294 (2019): 5261-5280.
- Jing D, Zhang Q, Yu H, et al. Identification of WISP1 as a novel oncogene in glioblastoma. Int J Oncol 51 (2017):1261-1270.
- Wang QY, Feng YJ, Ji R. High expression of WISP1 promotes metastasis and predicts poor prognosis in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci 24 (2020): 10445-10451.
- Chen YZ, Sun DQ, Zheng Y, et al. WISP1 silencing confers protection against epithelial-mesenchymal transition of renal tubular epithelial cells in rats via inactivation of the wnt/beta-catenin signaling pathway in uremia. J Cell Physiol 234 (2019): 9673-9686.
- Gaudreau PO, Clairefond S, Class CA, et al. WISP1 is associated to advanced disease, EMT and an inflamed tumor microenvironment in multiple solid tumors. Oncoimmunology 8 (2019): e1581545.
- Liao X, Bu Y, Xu Z, et al. WISP1 Predicts Clinical Prognosis and Is Associated With Tumor Purity, Immunocyte Infiltration, and Macrophage M2 Polarization in Pan-Cancer. Front Genet 11 (2020): 502.
- Tao W, Chu C, Zhou W, et al. Dual Role of WISP1 in maintaining glioma stem cells and tumor-supportive macrophages in glioblastoma. Nat Commun 11 (2020): 3015.
- Harb-de la Rosa A, Acker M, Kumar RA, et al. Epigenetics application in the diagnosis and treatment of bladder cancer. Can J Urol 22 (2015): 7947-7951.
- Hou G, Xu W, Jin Y, et al. MiRNA-217 accelerates the proliferation and migration of bladder cancer via inhibiting KMT2D. Biochem Biophys Res Commun 519 (2019): 747-753.
- Zhao X, Li D, Zhao ST, et al. MiRNA-616 aggravates the progression of bladder cancer by regulating cell proliferation, migration and apoptosis through downregulating SOX7. Eur Rev Med Pharmacol Sci 23 (2019): 9304-9312.
- Sekar D. miRNA 21: a novel biomarker in the treatment of bladder cancer. Biomark Med 14 (2020): 1065-1067.
- Yin XH, Jin YH, Cao Y, et al. Development of a 21-miRNA Signature Associated With the Prognosis of Patients With Bladder Cancer. Front Oncol 9 (2019): 729.
- Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 9 (2007): 166-180.
- Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 6 (2004): 1-6.
- Lanczky A, Nagy A, Bottai G, et al. miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res Treat 160 (2016): 439-446.
- Agarwal V, Bell GW, Nam JW, et al. Predicting effective microRNA target sites in mammalian mRNAs. Elife 4 (2015).
- Zhang LH, Wang Y, Fan QQ, et al. Up-regulated Wnt1-inducible signaling pathway protein 1 correlates with poor prognosis and drug resistance by reducing DNA repair in gastric cancer. World J Gastroenterol 25 (2019): 5814-5825.
- Lee HL, Chiou HL, Wang SS, et al. WISP1 genetic variants as predictors of tumor development with urothelial cell carcinoma. Urol Oncol 36 (2018): 15-21.
- Zheng Y, Chen CJ, Lin ZY, et al. Circ_KATNAL1 regulates prostate cancer cell growth and invasiveness through the miR-145-3p/WISP1 pathway. Biochem Cell Biol 98 (2020): 396-404.
- Mao A, Tang J, Tang D, et al. MicroRNA-29b-3p enhances radiosensitivity through modulating WISP1-mediated mitochondrial apoptosis in prostate cancer cells. J Cancer 11 (2020): 6356-64.
- Wang L, Sun J, Cao H. MicroRNA-384 regulates cell proliferation and apoptosis through directly targeting WISP1 in laryngeal cancer. J Cell Biochem 120 (2019): 3018-3026.
- Pommier A, Varilh J, Bleuse S, et al. miRNA repertoires of cystic fibrosis ex vivo models highlight miR-181a and miR-101 that regulate WISP1 expression. J Pathol 253 (2020): 186-197.
- Oto J, Plana E, Fernandez-Pardo A, et al. Identification of miR-29c-3p as a Robust Normalizer for Urine microRNA Studies in Bladder Cancer. Biomedicines 8 (2020): 447.
- Fang R, Huang Y, Xie J, et al. Downregulation of miR-29c-3p is associated with a poor prognosis in patients with laryngeal squamous cell carcinoma. Diagn Pathol 14 (2019): 109.
- Hu Z, Cai M, Zhang Y, et al. miR-29c-3p inhibits autophagy and cisplatin resistance in ovarian cancer by regulating FOXP1/ATG14 pathway. Cell Cycle 19 (2020): 193-206.
- Wang H, Fu L, Wei D, et al. MiR-29c-3p Suppresses the Migration, Invasion and Cell Cycle in Esophageal Carcinoma via CCNA2/p53 Axis. Front Bioeng Biotechnol 8 (2020): 75.
- Yu G, Zhou H, Xu K, et al. Mir-29c-3p targeting TUG1 affects migration and invasion of bladder cancer cells by regulating CAPN7 expression. Nan Fang Yi Ke Da Xue Xue Bao 40 (2020): 1325-1331.
- Liu Z, Zhang H, Sun L, et al. miR-29c-3p Increases Cell Viability and Suppresses Apoptosis by Regulating the TNFAIP1/NF-kappaB Signaling Pathway via TNFAIP1 in Abeta-Treated Neuroblastoma Cells. Neurochem Res 45 (2020): 2375-2384.


 Impact Factor: * 4.1
Impact Factor: * 4.1 Acceptance Rate: 74.74%
Acceptance Rate: 74.74%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 