Prognostic Value of Survival-Associated Splicing Factor SNRPA1 Overexpression and its Potential Mechanism in Liver Cancer
Mengying Liu1, Yueda Lu1, Lei Wei2, Xiaowo Wang2, Yinying Lu3, Xiaodong Jia3, Shihui Wang1, Wenlin An1,4*
1College of Life Science and Technology, Beijing University of Chemical Technology, Beijing, China
2Ministry of Education Key Laboratory of Bioinformatics, Center for Synthetic and Systems Biology, Bioinformatics Division, Beijing National Research Center for Information Science and Technology, Department of Automation, Tsinghua University, Beijing, China
3The comprehensive liver cancer center, Fifth medical center of Chinese PLA general hospital, Beijing, China
4National Vaccine and Serum Institute (NVSI), China National Biotech Group (CNBG), No. 38 Jing Hai Second Road, Beijing, 101111, China
*Corresponding Author: Wenlin An, College of Life Science and Technology, Beijing University of Chemical Technology, Beijing, China.
Received: 15 March 2022; Accepted: 01 April 2022; Published: 19 April 2022
Article Information
Citation:
Mengying Liu, Yueda Lu, Lei Wei, Xiaowo Wang, Yinying Lu, Xiaodong Jia, Shihui Wang, Wenlin An. Prognostic Value of Survival-Associated Splicing Factor SNRPA1 Overexpression and its Potential Mechanism in Liver Cancer. Journal of Cancer Science and Clinical Therapeutics 6 (2022): 188-206
View / Download Pdf Share at FacebookAbstract
Background: Small Nuclear Ribonucleoprotein Polypeptide A (SNRPA1) is a Splicing Factor (SF) responsible for the processing of pre-mRNA into mRNA. The expression level of SNRPA1 associated with several cancer types. However, the expression level of SNRPA1 and its role as a splicing factor in hepatocellular carcinoma remain unclear. The purpose of this study was to explore the clinicopathological characteristics and prognostic significance of SNRPA1 mRNA expression level and Percent-Spliced-In (PSI) values in liver cancer.
Methods: A total of 418 RNA−Seq and clinical data were downloaded from The Cancer Genome Atlas (TCGA) database. Alternative Splicing (AS) profiles were downloaded from TCGA SpliceSeq. Wilcoxon rank-sum tests were used to compare the expression levels of normal tissues with tumor tissues. The Kruskal Wallis tests were used to analyze the expression difference in grade, stage, and T classification among normal tissues with tumor tissues. The Kaplan-Meier analysis method was used to draw the survival curves. Univariate and multivariate Cox analyses were employed to estimate the prognostic value of SNRPA1. Gene Set Enrichment Analysis (GSEA) was performed to identify the signaling pathways. Then we used univariate and Pearson's correlation tests to analyze the correlation between SFs and Exon Skip (ES) events. Wilcoxon rank-sum tests were applied to analyze the relationships between different spliceosome and cancers. Furthermore, we evaluated the expression levels of SNRPA1 with clinical samples and The Clinical Proteomic Tumor Analysis Consortium (CPTAC) database.
Results: The level of SNRPA1 mRNA expression in liver cancer was significantly up-regulated in tumor tissues compared with normal tissues (p=1.411e−27) in liver cancer and was positively correlated with survival status (p=0
Keywords
<p>Liver cancer; Prognosis; SNRPA1; Splicing factors; SCP2</p>
Article Details
Abbreviations:
AS: Alternative Splicing; BRCA: Breast Invasive Carcinoma; CPTAC: The Clinical Proteomic Tumor Analysis Consortium; COAD: Colon Adenocarcinoma; ES: Exon Skip; FDR: False Discovery Rate; GO: Gene Ontology; GSEA: Gene Set Enrichment Analysis; KEGG: Kyoto Encyclopedia of Gene and Genomes; LIHC: Liver Hepatocellular Carcinoma; LUAD: lung Adenocarcinoma; OS: Overall Survival Rate; PSI: Percent−Spliced−In; RFS: Recurrence−Free Survival; SF: Splicing Factor; SNRPA1: Small Nuclear Ribonucleoprotein Polypeptide A; STAD: Stomach Adenocarcinoma; TCGA: The Cancer Genome Atlas
1. Introduction
Liver cancer is predicted to be the sixth most commonly diagnosed cancer and the fourth most common cause of cancer death worldwide in 2018, with about 841,000 new cases and 7,82,000 deaths each year [1]. Similar to other malignant tumors, the pathogenesis of liver cancer is very complicated. The factors contributing to the occurrence of liver cancer include chronic hepatitis virus infection, alcohol, drugs, and genetic factors [2]. So far, a surgical operation is still the first choice in treatment of liver cancer. It was reported that 5-year Recurrence Free Survival Rate (RFS) and 5-year Overall Survival Rates (OS) were only 30.8%-42.8% and 42.9%-60, respectively [3, 4]. Even in the early stage of liver cancer, its 5-year cumulative recurrence rate was reported to be as high as 57.2%, and the 5-year overall survival rate was only 76.4% [5]. Therefore, it is a clinically important to identify a reliable biomarker that can be used for diagnosis and to predict the prognosis of liver cancer.
Multiple studies have shown that specific Alternative Splicing (AS) events such as cell proliferation, angiogenesis, tumor metastasis, and immune escape, are associated with the development and progression of cancer [6, 7]. There are seven common patterns of AS events: Alternate Acceptor Site (AA), Alternate Donor site (AD), Alternate Promoter (AP), Alternate Terminator (AT), Exon Skip (ES), Mutually Exclusive Exons (ME), and Retained Intron (RI) [8]. More importantly, the expression of Specific Splicing Factors (SFs) could regulate AS events [9, 10].
Oncofetal splicing factor MBNL3 could promote tumorigenesis and indicates poor prognosis of hepatocellular carcinoma patients. The knockdown of MBNL3 almost completely abolishes hepatocellular carcinoma tumorigenesis [11]. Therefore, understanding the roles of splicing factors and splicing events during tumorigenesis would open new avenues for targeted therapies.
Small nuclear ribonucleoprotein polypeptide A (SNRPA1) is a spliceosome component responsible for the processing of pre−mRNA into mRNA. It is a necessary factor for male reproductive ability and the defects of spliceosome could affect the differentiation of human spermatogonia [12]. SNRPA1 regulates the expression of CDK1, PIK3R1, VEGFC, and MKI67 in colorectal cancer. It can be recruited to laser−induced DNA damage sites to prevent R−loop−induced DNA damage [13, 14]. In another study, SNRPA1 and TCF7L2 were found to bind to the insertion allele of rs386772267, a genetic insertion which is associated with the increased risk of pancreatic cancer [15]. SNRPA1 could interact with certain structural splicing enhancer, which is enriched near cassette exons with increased inclusion in highly metastatic cells of breast cancer to promote cassette exon inclusion. This interaction enhances metastatic lung colonization and cancer cell invasion [16].
Although several studies have reported this gene [13-16], the prognostic value and potential mechanism of SNRPA1 as a splicing factor in liver cancer remain unclear. As a SF associated with hepatocellular carcinoma, it will be important to understand the function and regulatory genes of SNRPA1. In this study we explored the SNRPA1 expression in liver cancer and the related signaling pathways through the Gene Set Enrichment Analysis (GSEA). We found that the main pathway of KEGG converged at a spliceosome. Pearson's correlation test indicated that SNRPA1 could modify the exon skip of SCP2. Further analysis revealed that SNRPA1 and SCP2 PSI values are significantly increased in many cancer types, suggesting that a particular type of splice is actively involved in cancer. The PSI values of these two genes were used to predict survival more accurately than the mRNA expression levels of SNRPA1 and SCP2. In brief, our analysis reveals the importance of SNRPA1 in tumorigenesis as a splicing factor in liver cancer.
2. Methods
2.1. Data acquisition and preprocessing
The Level 3 expression data were downloaded from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/repository). SNRPA1 expression pattern and its prognostic significance were validated from liver cancer tissues paired with normal liver tissues. We analyzed the AS profiles via TCGA SpliceSeq (http://bioinformatics.mdanderson.org/TCGASpliceSeq), a resource for investigating of cross-tumor and tumor-normal alterations in mRNA splicing patterns of RNA-Seq data [17]. The percent-spliced-in (PSI) value of the database indicates the percentage of a transcript element over the total normalized reads for that event with values ranging from 0 to 1 [18]. For validation, we downloaded the datasets from The NCI Clinical Proteomic Tumor Analysis Consortium(CPTAC)(https://proteomics.cancer.gov/programs/cptac), aiming at characterizing the protein datasets in tumors to further confirm our findings [19].
2.2. Clinical sample acquisition
4 paired of tumor and adjacent non-tumor liver tissues were obtained from the Fifth medical center of Chinese PLA general hospital. All the patients underwent primary curative resection and received no prior anticancer treatments. Tissue samples were collected within 30 min after operation and snap-frozen in liquid nitrogen. All experiments were performed in accordance with relevant guidelines and regulations. The study was approved by Fifth medical center of Chinese PLA general hospital, and written informed consent was obtained from each patient.
2.3 Statistical analysis
2.3.1. SNRPA1 expression and clinical analysis
Statistical analyses were performed in R (version 3.6.3) software using R packages: limma, beeswarm, survival, and survminer [20-24]. The Wilcoxon rank-sum tests were applied to compare the relative expression level of normal tissues with tumor tissues. The Kruskal Wallis tests were used to analyze the expression difference in grade, stage, and T classification among normal tissues with tumor tissues. Then tumor tissues were divided into low and high expression groups by using SNRPA1 expression median level as a cut-off point. Overall survival rates (follow-up time >30 days) were compared between the high and low expression groups via Kaplan−Meier analysis. The independent prognostic value of SNRPA1 expression on liver cancer was assessed by univariate and multivariate Cox analyses. p<0.05 was considered statistically significant.
2.3.2. Gene set enrichment analysis of SNRPA1
Gene Set Enrichment Analysis (GSEA) was performed to identify the signaling pathways related to the regulatory mechanism of SNRPA1 by using the GSEA v4.0.3 software [25]. Kyoto Encyclopedia of Gene and Genomes (KEGG) gene sets (c2.cp.kegg.v7.1.symbols.gmt) and Gene Ontology (GO) gene sets (c5.all.v7.1.symbols.gmt) from the Molecular Signatures Database (MSigDB) (http://www.broad.mit.edu/gsea/msigdb/index.jsp) were utilized to analyze pathways. The tumor tissues were divided into low and high expression groups using SNRPA1 expression median level as a cut-off point. Data sets with a p <0.05 and a false discovery rate (FDR) <0.25 were considered to be significantly enriched.
2.3.3. Analysis of the role of SNRPA1 as a splicing factor
The ‘upset’ function in the ‘UpSet’ R package was used to visualize the interactive AS events between the seven AS types, to clearly show quantitative results of multiple interactive sets. The prognostic relationship between the PSI value of AS events and overall survival rates (follow-up time >30 days) were performed using the univariate Cox proportional hazards regression model.
2.3.4. Correlation analysis of AS events and SFs
A total of 404 splicing factors were retrieved from the SpliceAid2 database [26]. Pearson correlation analysis was performed to explore the interaction and correlation between SFs and significant AS events (p<0.05, OS−related ASs). The screening conditions were a correlation coefficient >0.6 or <−0.6, and a p <0.001. Finally, we visualized the regulatory networks between SFs and Exon Skip (ES) events using Cytoscape (version 3.7.2) [27].
2.3.5. Relationships between different splices and cancers
For further analysis of the relationships between different splices and cancers, we downloaded the genes of interest in different cancers on the TCGA SpliceSeq database. They included Breast Invasive Carcinoma (BRCA), colon adenocarcinoma (COAD), Liver Hepatocellular Carcinoma (LIHC), Lung Adenocarcinoma (LUAD), and Stomach Adenocarcinoma (STAD). Then the Wilcoxon rank-sum tests were applied to compare the expression level of normal tissues with cancer tissues in different cancers.
2.3.6. The relationships between PSI values of SNRPA1, SCP2 and clinical analysis
We used the PSI values of SNRPA1 and SCP2 to analyze the differences in grade, stage, and T classification, among subgroups via the Kruskal Wallis tests. We then drew the survival curves according to PSI value (median level as a cut-off point) via Kaplan−Meier analysis (follow-up time >30 days).
2.3.7. SNRPA1 expression in CPTAC database
To find out whether the genes identified from the TCGA database also are of prognostic significance in protein level, we downloaded and analyzed mass spectrometry-based proteomics data from CPTAC database. The Wilcoxon rank-sum tests were applied to compare the protein expression level of normal tissues with tumor tissues. Overall survival rates (follow−up time >30 days) were compared between the high and low expression groups via Kaplan−Meier analysis. p <0.05 was considered statistically significant.
2.4. Dection of protein expression levels of SNRPA1 in clinical samples
We analyzed the proteomics of 4 pairs of tumor tissues and its adjacent non-tumor liver tissues through 4D Label-free detection techniques: TIMS-TOF Pro mass spectrometry (MS/MS). The results were processed using Maxquant search engine (v1.6.6.0). Retrieval arguments is setting to Homo sapiens 9606 SP 20191115 (20380 sequences). Tandem mass spectra were searched against Human Uniprot database concatenated with reverse decoy database.
3. Results
3.1. SNRPA1 expression and clinical analysis
A set of data from 418 patients were downloaded from the TCGA database with corresponding patient demographic and clinical characteristics data including age, gender, histological grade, stage, T/N/M classification and survival status of liver cancer (Table 1). Paired tumor, adjacent non-tumor liver tissues from a cohort of 316 HBV-related HCC patients were downloaded from the current Clinical Proteomic Tumor Analysis Consortium (CPTAC) project [28]. SNRPA1 mRNA expression level was significantly up−regulated in tumor tissues compared with normal tissues (p=1.411e−27, Figure 1A) in liver cancer. A paired comparison between normal and liver cancer tissue from the same patients also showed a significant up−regulation (p=4.08e−16, Figure 1B). SNRPA1 expression level showed a positive correlation with survival status (p=0.035, Figure 1C). The raw data of the survival analysis were shown in Supplementary Table S1. Furthermore, significant differences were observed in SNRPA1 expression based on histological grade and T classification (Figure 1D-1F). Univariate and multivariate Cox analyses indicated that the SNRPA1 mRNA expression (hazard ratio [HR]=1.08, 95% confidence interval [CI]: 1.02–1.14, p=0.005, Table 2) could be a useful biomarker for liver cancer prognosis.
|
Characteristics |
Number of patients (%) |
|
|
Age |
<55 |
120(28.71) |
|
≥55 |
256(61.24) |
|
|
Not available |
42(10.05) |
|
|
Gender |
Female |
146(34.93) |
|
Male |
254(60.77) |
|
|
Not available |
18(4.31) |
|
|
Histological grade |
G1 |
55(13.16) |
|
G2 |
180(43.06) |
|
|
G3 |
124(29.67) |
|
|
G4 |
13(3.11) |
|
|
Not available |
46(11) |
|
|
Stage |
I |
194(46.41) |
|
II |
98(23.44) |
|
|
III |
90(21.53) |
|
|
IV |
12(2.87) |
|
|
Not available |
24(5.74) |
|
|
T Classification |
T1 |
204(48.8) |
|
T2 |
107(25.6) |
|
|
T3 |
90(21.53) |
|
|
T4 |
14(3.35) |
|
|
TX |
1(0.24) |
|
|
Not available |
2(0.48) |
|
|
M Classification |
M0 |
303(72.49) |
|
M1 |
8(1.91) |
|
|
MX |
107(25.6) |
|
|
N Classification |
N0 |
290(69.38) |
|
N1 |
8(1.91) |
|
|
NX |
119(28.47) |
|
|
Not available |
1(0.24) |
|
|
Survival status |
Death |
147(35.17) |
|
Survival |
271(64.83) |
|
|
Not available data and TX data were not used for subsequent analysis |
||
Table 1: Clinical characteristics of the liver cancer patients.
|
Univariate analysis |
Multivariate analysis |
|||||
|
Parameters |
HR |
95%CI |
P-value |
HR |
95%CI |
P-value |
|
age |
1.01 |
1–1.03 |
0.177 |
|||
|
gender |
0.82 |
0.56–1.21 |
0.317 |
|||
|
grade |
1.12 |
0.87–1.45 |
0.382 |
|||
|
stage |
1.67 |
1.36–2.06 |
0 |
1.19 |
0.51–2.8 |
0.68 |
|
T classification |
1.65 |
1.36–2.01 |
0 |
1.41 |
0.63–3.18 |
0.402 |
|
SNRPA1 |
1.08 |
1.03–1.14 |
0.002 |
1.08 |
1.02–1.14 |
0.005 |
|
P-values in Bold indicate p <0.05. HR: hazard ratio; CI: confidence interval. |
||||||
Table 2: Univar ate analysis and multivariate analyses of the correlation between SNRPA1 expression and clinical parameters.
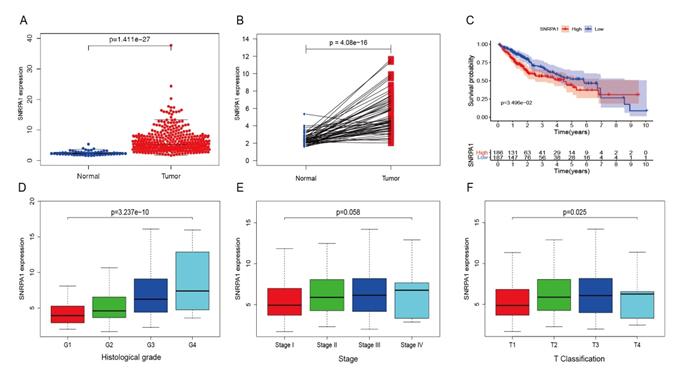
Figure 1: SNRPA1 expression in liver cancer. Comparison of SNRPA1 mRNA expression between (A) normal and cancer tissues and (B) paired samples. (C) Kaplan−Meier curves for OS in patients. Comparison of SNRPA1 expression according to clinical parameters: histologic grade (D), stage (E), and T classification (F).
3.2. SNRPA1 gene set enrichment analysis in liver cancer
To identify the potential mechanisms of SNRPA1 expression on liver cancer prognosis, we conducted the GSEA (GO and KEGG pathway enrichment analysis) between low and high SNRPA1 expression groups (Table 3). The GO and KEGG analyses results showed processes and pathways associated with AS. “RNA splicing”, “small nuclear ribonucleoprotein complex”, “RNA splicing via transesterification reactions”, “spliceosomal complex” and “mRNA processing” were enriched in GO analysis. “Spliceosome”, as well as some carcinogenesis and development associated pathways, like “DNA replication”, “base excision repair”, “RNA degradation” and “cell cycle” were enriched in KEGG analysis. These related results have been shown in Figure 2. Our results suggested that SNRPA1 is related to other gene functions through alternative splicing.
Table 3: The top 10 enriched GO and KEGG pathways of high SNRPA1 expression groups.
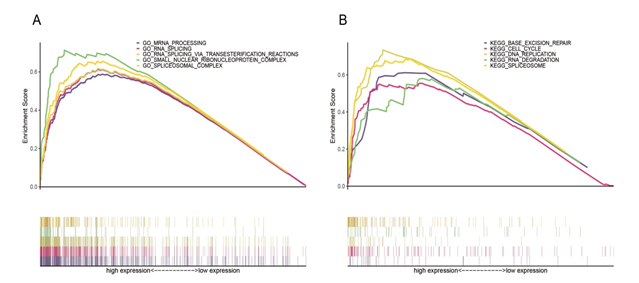
Figure 2: The main enriched GO and KEGG pathways of high SNRPA1 expression sets. The SNRPA1 expression groups of GO (A) and KEGG pathways (B) analyses.
3.3. Alternative splicing profiles of liver cancer in TCGA
As SNRPA1 plays a role in alternative splicing, we analyzed the occurrence of AS events in the TCGA database. By analyzing AS events of 418 cases of liver cancer patients from TCGA, we found 2666 AAs in 1937 genes, 2331 ADs in 1663 genes, 6325 APs in 2566 genes, 8087 ATs in 3532 genes, 12327 ESs in 5331 genes, 137 MEs in 135 genes, and 2263 RIs in 1561 genes. Detailed information about the specific AS types of genes was visualized in the Upset plot (Figure 3A).
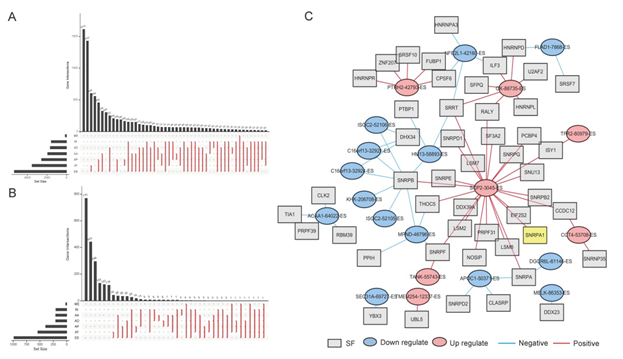
Figure 3: General characteristics of AS and OS-related AS events. (A) The UpSet plot for seven interactions types of AS events in liver cancer, one gene may have up to 6 types of AS. (B) UpSet plot for significant OS−related AS types. (C) The related genes interaction networks of ES events. AA: alternate acceptor site; AD: alternate donor site; AP: alternate promoter; AT: alternate terminator; ES: exon skip; ME: mutually exclusive exons; RI: retained intron.
3.4. Analysis of OS−related AS events with univariate Cox
AS events data was used to perform univariate analyses for OS. The results of the univariate Cox proportional hazards regression are shown in Supplementary Table S2. Inclusively, 222 AAs, 226 ADs, 618 APs, 891 ATs, 1272 ESs, 16 MEs, and 221 RIs were significantly altered (p <0.05). The Upset plot of significant OS−related AS types was shown in Figure 3B. The results shown that SNRPA1 (SNRPA1–32758–ES), belonging to ES events, was significantly (p=0.003) associated with OS in the univariate Cox model (Hazard Ratio [HR]=155.76, 95% Confidence Interval [CI]: 5.26 – 4612.99). The PSI value of every patient has been shown in Supplementary Table S3.
3.5. ES events-related genes interaction networks construction
With access to RNA-seq data and corresponding clinical information of liver cancer patients, we identified 404 candidate SFs whose expression levels were significantly associated with OS related ASs events. Pearson correlation analysis was performed to explore the interaction and correlation between SFs and significant AS events. The results were shown that SNRPA1 (COR=0.606, p=7.18E-34), as well as LSM8, PCBP4, LSM2, SNRPB2, NOSIP, SNRPG, SNRPE, SNRPF, SF3A2, PRPF31, EIF2S2, SNRPA, DDX39A, SNRPD1, THOC5, LSM7, ISY1, SRRT, SNU13, RALY, CCDC12, and SNRPB were related to SCP2 (SCP2-3045-ES) (Table 4). We constructed networks between the prognosis associated ES events and survival associated SFs to identify the underlying interactions (Figure 3C). A total of 48 SFs and 21 AS events were constructed. Ultimately, 69 nodes and 72 edges were established in the PPI networks, which included 14 down-regulated AS events and 7 up-regulated AS events. The SFs and AS events that correlated positively (COR >0.6) and negatively (COR <−0.6) were shown by red and blue edges, respectively. From the results, SCP2 could be regulated by 23 SFs, and one of them is SNRPA1.
|
SF |
AS |
COR |
p−value |
|
LSM8 |
SCP2−3045−ES |
0.64622 |
1.09E−39 |
|
PCBP4 |
SCP2−3045−ES |
0.606735 |
5.80E−34 |
|
LSM2 |
SCP2−3045−ES |
0.671344 |
8.47E−44 |
|
SNRPB2 |
SCP2−3045−ES |
0.621021 |
6.10E−36 |
|
NOSIP |
SCP2−3045−ES |
0.62959 |
3.54E−37 |
|
SNRPG |
SCP2−3045−ES |
0.633561 |
9.18E−38 |
|
SNRPE |
SCP2−3045−ES |
0.600225 |
4.29E−33 |
|
SNRPF |
SCP2−3045−ES |
0.702603 |
1.64E−49 |
|
SF3A2 |
SCP2−3045−ES |
0.626232 |
1.09E−36 |
|
PRPF31 |
SCP2−3045−ES |
0.630417 |
2.68E−37 |
|
SNRPA1 |
SCP2−3045−ES |
0.606052 |
7.18E−34 |
|
EIF2S2 |
SCP2−3045−ES |
0.655976 |
3.07E−41 |
|
SNRPA |
SCP2−3045−ES |
0.692363 |
1.47E−47 |
|
DDX39A |
SCP2−3045−ES |
0.641328 |
6.19E−39 |
|
SNRPD1 |
SCP2−3045−ES |
0.716852 |
2.28E−52 |
|
THOC5 |
SCP2−3045−ES |
0.604718 |
1.08E−33 |
|
LSM7 |
SCP2−3045−ES |
0.711111 |
3.39E−51 |
|
ISY1 |
SCP2−3045−ES |
0.748097 |
2.66E−59 |
|
SRRT |
SCP2−3045−ES |
0.633006 |
1.11E−37 |
|
SNU13 |
SCP2−3045−ES |
0.724853 |
4.71E−54 |
|
RALY |
SCP2−3045−ES |
0.666096 |
6.60E−43 |
|
CCDC12 |
SCP2−3045−ES |
0.619867 |
8.89E−36 |
|
SNRPB |
SCP2−3045−ES |
0.734261 |
4.13E−56 |
|
COR: correlation coefficient |
|||
Table 4: The correlation analysis of AS events and the expression of SFs for SCP2.
3.6. Relationships between different splices and cancer types
Based on the results in 3.5, SNRPA1 as a splicing factor could has the correlation with the AS events of SCP2, we wanted to figure out what kind of SNRPA1/SCP2 splices are the most associated with different cancer types. The different splices have been shown in Figure 4A. The results of comparison with PSI values of normal and tumor tissues of liver cancer showed that SNRPA1 exon 6 skip (SNRPA1_exon_6) and SCP2 exon 12 skip (SCP2_exon_12) correlated in liver cancer (Figure 4B). The PSI values (the following refers to the PSI value of the SNRPA1 exon 6 skip or the SCP2 exon 12 skip) of tumor tissues were significantly different from the PSI values of normal tissues with Wilcoxon rank-sum tests in LICH (p <0.001). Further analyses in BRCA, COAD, LUAD and STAD showed the PSI values of SNRPA1_exon_6 and SCP2_exon_12 were significantly increasing in tumor tissues (p <0.001 for each cancer type, Figure 4C−4D). The raw data of PSI values and related cancer types were shown in Supplementary 4.
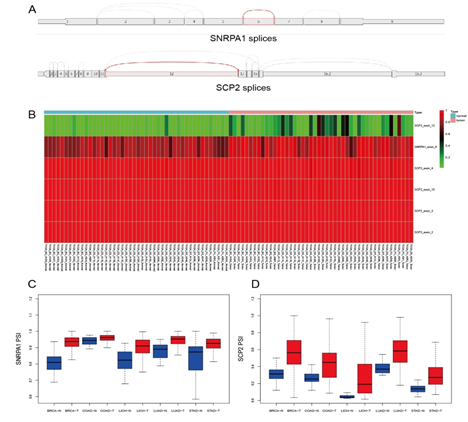
Figure 4: Relationships between different spliceosome and cancers. (A) The SNRPA1 and SCP2 splices. (B) Heatmap of the different spliceosomes in LICH (C) SNRPA1 exon 6 skip (SNRPA1_exon_6) PSI value in different cancers. (D) SCP2 exon 12 skip (SCP2_exon_12) PSI value in different cancers. BRCA: breast invasive carcinoma; COAD: colon adenocarcinoma; LIHC: liver hepatocellular carcinoma; LUAD: lung adenocarcinoma; STAD: stomach adenocarcinoma; T: tumor tissues; N: normal tissues.
3.7. The relationships between PSI values of SNRPA1, SCP2 and clinical analysis
Further analysis showed that the PSI values of the two genes were associated with survival status, histological grade, and T classification. SNRPA1 and SCP2 PSI values showed the positive correlations with survival status (p=3.022e-04 and p=2.932e−03, respectively, Figure 5A-B). The raw data of the survival analysis were shown in Supplementary Table S1. Furthermore, the PSI values of SNRPA1 and SCP2 were significant different among subgroups in histological grade (p=3.87e−07 and p=1.598e−08), stage (p >0.005 and p=0.007), and T classification (p=0.003 and p=0.002) (Figure 5C-H).
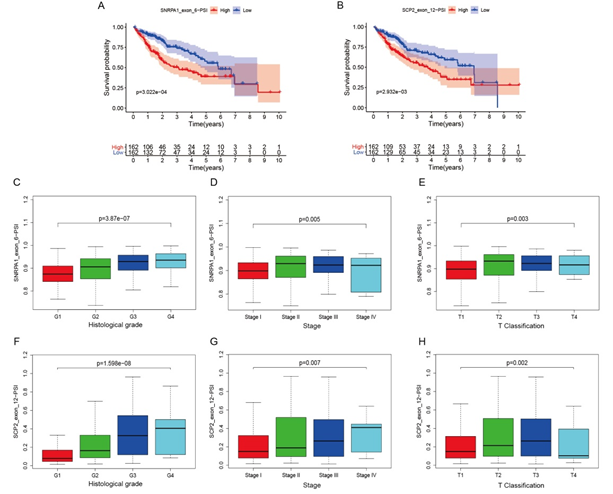
Figure 5: The relationships between PSI values of SNRPA1, SCP2 and clinical analysis. The high and low PSI value groups of SNRPA1 and SCP2 corresponded with Kaplan−Meier curves for OS in patients (A-B). PSI values of SNRPA1 and SCP2 were compared with clinical parameters: histologic grade, stage, and T classification (C-H).
3.8. CPTAC database and experiment validation the SNRPA1 expression at protein level
For the CPTAC analyzed results, SNRPA1 protein expression level was significantly up−regulated in tumor tissues compared with normal tissues (p=3.197e-47, Figure 6A) in liver cancer. Our clinical samples results were consistent with the CPTAC database. SNRPA1 protein expression level was significantly up−regulated in tumor tissues compared with normal tissues (p=0.029, Figure 6B).
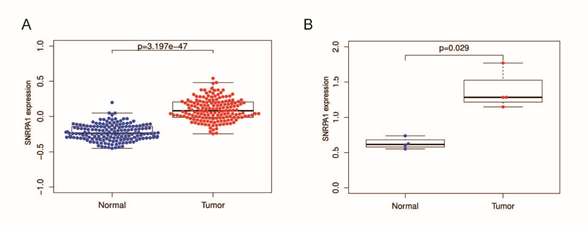
Figure 6: SNRPA1 expression in protein level. (A) Comparison of SNRPA1 protein expression between normal and cancer tissues in CPTAC database. (B) Comparison of SNRPA1 protein expression between normal and cancer tissues in 4 pairs of clinical samples.
4. Discussion
Liver cancer is one of the most common cancers worldwide. Although significant improvement in diagnosis and treatment has been witnessed in clinical practice, the prognosis of liver cancer remains considerably unfavorable [29]. In recent years, bioinformatics tools have been employed to screen for molecular markers. Identification of such new molecular markers is likely to improve the prognosis and survival rate of liver cancer patients. In our study, we found that high expression of SNRPA1 was associated with histological grade, T classification, and poor survival status in liver cancer. The validation of CPTAC database and our clincal samples also supported this point of view. Univariate and multivariate Cox analyses indicated that the SNRPA1 mRNA expression level might be a useful biomarker for predicting the prognosis of liver cancer. The enriched GO and KEGG pathway analyses performed using GESA method showed the SNRPA1 is a spliceosome. SNRPA1 is a splicing factor which is responsible for the processing of pre−mRNA into mRNA to act on certain specific AS events. We combined clinical information and AS events in this study to analyze the AS events associated with survival in liver cancer. The univariate Cox analysis result showed that SNRPA1 of ES events could be an independent prognostic factor. Pearson's correlation test indicated that SNRPA1 could modify the exon skip of SCP2. Sterol carrier protein 2 (SCP2) is well recognized as an intracellular cholesterol trafficking protein that targets cholesterol to cholesterol−rich membrane microstructural domains [30]. Different splicing patterns in one gene produce diverse isoforms. For this reason, the regulation and mechanisms of AS are highly complex in cancer [31]. Further excavation of the relationship between clinical information and spliceosomes revealed that the PSI values of SNRPA1 exon 6 skip and SCP2 exon 12 skip correlated with LICH, BRCA, COAD, LUAD and STAD. Further analysis revealed that PSI values could have a positive correlation with survival status and portrayed were significant differences among subgroups in histological grade, cancer stage as well as T classification.
Overall, we integrated clinical information with AS events to reveal the underlying mechanism of SNRPA1. Based on our results, SNRPA1 is a potential prognostic marker for liver cancer and may play a role in exon skip of SCP2. The PSI values of SNRPA1 exon 6 skip and SCP2 exon 12 skip were significantly increased in many cancer types, suggesting that a particular type of splices is actively involved in cancer. Furthermore, the use of PSI values of SNRPA1 to predict survival status (p=3.022e−04) is significantly better than the use of mRNA expression of this gene (p=0.035). In conclusion, we have provided comprehensive landscape of splicing factor SNRPA1 in patients with liver cancer and identified OS-associated AS events. It might be potentially application value in clinical practice. In addition, we demonstrated that the prognosis-related AS events could be applied to build predictive factors with high accuracy to stratify survival risk compared to mRNA expression level in liver cancer. Deep-mining analysis of AS patterns and SF might indeed show new oncological drivers and confer some potential insights into carcinogenesis mechanism.
Declarations
Ethics approval and consent to participate
All experiments were performed in accordance with relevant guidelines and regulations. The study for using human tissues was approved by the Ethics Review Committee of Fifth Medical Center of Chinese PLA General Hospital, and written informed consent was obtained from each patient.
Consent for publication
Consent for publication not applicable in this article.
Availability of data and materials
The datasets used or analysed during the current study are available from the TCGA database (https://portal.gdc.cancer.gov/repository) and TCGA SpliceSeq(http://bioinformatics.mdanderson.org/TCGASpliceSeq). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD025790. All datasets also could be obtained from the corresponding authors.
Competing Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Author Contributions
WA supervised and participated in the design of this study. ML conceived, designed and carried out the study, and wrote the initial manuscript. ML and YL carried out literature search, data acquisition and analyses. YL collected and provided important background information. LW, XW, YL, XJ and SW provided assistance for data analyses and statistical analyses. WA and LW edited, corrected and proofread the full contents of the paper. All authors read and approved the final manuscript.
Funding
This work was supported by National Key R & D Program "Synthetic Biology" key special project (2018YFA0903000) foundation with the samples testing and the layout fees; and by National Science and Technology Major Project during the 13th Five-Year Plan Period (2019ZX09721001-007) foundation with the article polishing fee; and by National Natural Science Foundation General Project (81672001), National Science Foundation of China (81672467) foundations with sample processing costs; and by Medical Big Data and AI R & D Project of General Hospital (2019MBD−025) foundation with researchers labor expenses.
Acknowledgments
We acknowledge the funding by National Science and Technology Major Project during the 13th Five-Year Plan Period, National Key R & D Program "Synthetic Biology" key special project, National Natural Science Foundation General Project, National Science Foundation of China, and Medical Big Data and AI R & D Project of General Hospital.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68 (2018): 394-424.
- Shariff MI, Cox IJ, Gomaa AI, et al. Hepatocellular carcinoma: current trends in worldwide epidemiology, risk factors, diagnosis and therapeutics. Expert Rev Gastroenterol Hepatol 3(2009): 353-67.
- Lazzara C, Navarra G, Lazzara S, et al. Does the margin width influence recurrence rate in liver surgery for hepatocellular carcinoma smaller than 5 cm? Eur Rev Med Pharmacol Sci 21 (2017): 523-529.
- Li C, Wen TF, Yan LN, et al. Liver resection versus liver resection plus TACE for patients with hepatocellular carcinoma beyond Milan criteria. J Surg Res 209 (2017): 8-16.
- Margonis GA, Sasaki K, Andreatos N, et al. Prognostic impact of complications after resection of early stage hepatocellular carcinoma. J Surg Oncol 115 (2017): 791-804.
- Munkley J, Livermore K, Rajan P, et al. RNA splicing and splicing regulator changes in prostate cancer pathology. Hum Genet 136 (2017): 1143-1154.
- Oltean S, Bates DO. Hallmarks of alternative splicing in cancer. Oncogene 33 (2014): 5311-5318.
- Ryan MC, Cleland J, Kim R, et al. SpliceSeq: a resource for analysis and visualization of RNA-Seq data on alternative splicing and its functional impacts. Bioinformatics (Oxford, England) 28 (2012): 2385-2387.
- Koh CM, Bezzi M, Low DH, et al. MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature 523 (2015): 96-100.
- Shilo A, Ben Hur V, Denichenko P, et al. Splicing factor hnRNP A2 activates the Ras-MAPK-ERK pathway by controlling A-Raf splicing in hepatocellular carcinoma development. RNA 20 (2014): 505-515.
- Yuan JH, Liu XN, Wang TT, et al. The MBNL3 splicing factor promotes hepatocellular carcinoma by increasing PXN expression through the alternative splicing of lncRNA-PXN-AS1. Nature cell biology 19 (2017): 820-832.
- Wu H, Sun L, Wen Y, et al. Major spliceosome defects cause male infertility and are associated with nonobstructive azoospermia in humans. Proc Natl Acad Sci USA 113 (2016): 4134-4139.
- Zeng Q, Lei F, Chang Y, et al. An oncogenic gene, SNRPA1, regulates PIK3R1, VEGFC, MKI67, CDK1 and other genes in colorectal cancer. Biomed Pharmacother 117 (2019): 109076.
- Tanikawa M, Sanjiv K, Helleday T, et al. The spliceosome U2 snRNP factors promote genome stability through distinct mechanisms; transcription of repair factors and R-loop processing. Oncogenesis 5(2016): 280.
- Hoskins JW, Ibrahim A, Emmanuel MA, et al. Functional characterization of a chr13q22.1 pancreatic cancer risk locus reveals long-range interaction and allele-specific effects on DIS3 expression. Hum Mol Genet 25 (2016): 4726-4738.
- Fish L, Khoroshkin M, Navickas A, et al. A prometastatic splicing program regulated by SNRPA1 interactions with structured RNA elements. Science (New York, NY) 372 (2021).
- Ryan M, Wong WC, Brown R, et al. TCGASpliceSeq a compendium of alternative mRNA splicing in cancer. Nucleic Acids Res 44 (2016): 1018-1022.
- Schafer S, Miao K, Benson CC, et al. Alternative Splicing Signatures in RNA-seq Data: Percent Spliced in (PSI). Curr Protoc Hum Genet 87 (2015):1161-116 4.
- Rudnick PA, Markey SP, Roth J, et al. A Description of the Clinical Proteomic Tumor Analysis Consortium (CPTAC) Common Data Analysis Pipeline. Journal of proteome research 15 (2016): 1023-1032.
- Team RDCJC. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria 14 (2009): 12-21.
- Matthew E, Ritchie BP, Di Wu, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research 43 (2015): 47.
- Eklund A. beeswarm: The Bee Swarm Plot, an Alternative to Stripchart (2016).
- Therneau T. A Package for Survival Analysis in S (2015).
- Alboukadel Kassambara, Marcin Kosinski, Biecek P. survminer: Drawing Survival Curves using 'ggplot2' (2019).
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102 (2005): 15545-15550.
- Piva F, Giulietti M, Nocchi L, et al. SpliceAid: a database of experimental RNA target motifs bound by splicing proteins in humans. Bioinformatics (Oxford, England) 25 (2009): 1211-1213.
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research 13 (2003): 2498-2504.
- Gao Q, Zhu H, Dong L, et al. Integrated Proteogenomic Characterization of HBV-Related Hepatocellular Carcinoma. Cell 179 (2019): 561-577.
- Altekruse SF, Henley SJ, Cucinelli JE, et al. Changing hepatocellular carcinoma incidence and liver cancer mortality rates in the United States. Am J Gastroenterol 109 (2014): 542-553.
- Burgardt NI, Gianotti AR, Ferreyra RG, et al. A structural appraisal of sterol carrier protein 2. Biochim Biophys Acta Proteins Proteom 1865 (2017): 565-577.
- Wang H, Zhang CZ, Lu SX, et al. A Coiled-Coil Domain Containing 50 Splice Variant Is Modulated by Serine/Arginine-Rich Splicing Factor 3 and Promotes Hepatocellular Carcinoma in Mice by the Ras Signaling Pathway. Hepatology 69 (2019): 179-195.


 Impact Factor: * 4.1
Impact Factor: * 4.1 Acceptance Rate: 74.74%
Acceptance Rate: 74.74%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 