The Mechanism of Hepatitis B Virus X Gene in Promoting Hepatocellular Carcinoma
Xinyu Zhou1, Donghong Liu2, Zishuai Li1, Jun Zhao2, Shiliang Cai1, Guangwen Cao1*
1Department of Epidemiology, Second Military Medical University, Shanghai, China
2The Third Affiliated Hospital of Second Military Medical University, Shanghai, China
*Corresponding Author: Guangwen Cao, Department of Epidemiology, Second Military Medical University, 800 Xiangyin Rd., Shanghai 200433, P. R. China
Received: 18 April 2022; Accepted: 13 May 2022; Published: 25 May 2022
Article Information
Citation: Xinyu Zhou, Donghong Liu, Zishuai Li, Jun Zhao, Shiliang Cai, Guangwen Cao. The Mechanism of Hepatitis B Virus X Gene in Promoting Hepatocellular Carcinoma. Journal of Cancer Science and Clinical Therapeutics 6 (2022): 222-233.
View / Download Pdf Share at FacebookAbstract
Primary liver cancer (PLC) was the third leading cause of cancer death worldwide and hepatocellular carcinoma (HCC) accounts for 75-85% of PLC cases. Chronic hepatitis B virus (HBV) infection is the major cause of HCC globally. HBV carcinogenesis depends on three factors: viral replication, integration and evolution, and HBV X gene (HBx) plays a major role in these processes. HBx determines HBV replication and is also the main viral gene for integration and evolution. Recently, numerous new carcinogenic mechanisms of HBx, including epigenetic modification, stem-like signal pathway, metabolic regulation, immune suppression, and drug resistance, have being continuously explored. This article reviews the mechanism of HBx and its mutation in the occurrence and development of HCC, in order to provide a reference for a comprehensive understanding of HBV-HCC.
Keywords
<p>Hepatitis B Virus; HBx; Mutation; Hepatocellular Carcinoma; Mechanism</p>
Article Details
1. Introduction
Primary liver cancer (PLC) was the sixth most commonly
diagnosed cancer and the third leading cause of cancer death worldwide in 2020 [1]. Hepatocellular carcinoma (HCC) comprises 75%-85% of PLC [1]. PLC is the leading cause of immature death (death before the mean life span of a given population) in China, among which HCC accounts for 94.6%. Chronic hepatitis B virus (HBV) infection accounts for 87.5% of HCCs in Eastern China [2-4]. HBV-related HCC (HBV-HCC) is associated with 10-year earlier onset, higher α-fetoprotein (AFP), and more microvascular invasion than HCC caused by other causes, indicating that HBV is more powerful in promoting HCC development than other etiological factors [4].
HBV replication, integration, and evolution are the main factors in the occurrence and development of HBV-HCC. HBx (HBV X) is a 17 kDa protein expressed from the X open reading frame (ORF) of HBV, with little sequence homology to any known genes, hence the name “X” [5]. HBx is a multifunctional factor that can regulate the HBV replication and activate cancer-promoting signal pathways [6]. During HBV-induced hepatocarcinogenesis, HBV typically adapts to the inflammatory microenvironment by integrating into the human genome and accumulating mutations [7]. HBx C-terminal truncation (Ct-HBx) resulting from HBV integration has been suggested to impact the development of HCC. HBx mutants, generated and accumulated in the chronic inflammation caused by HBV, play a complicated role in HCC [8]. Therefore, understanding the mechanism of HBx and its mutants in HCC can help understand the pathogenesis of HBV-HCC and provide novel prophylactic and therapeutic options for HBV-HCC.
2. Features of HBx Gene and HBx Protein
In the HBV genome, there are four overlapping ORFs, namely ORF-P, -S, -C, and –X [9]. The four ORFs encoding seven proteins (pre-S1, pre-S2, S, pre-C, C, viral polymerase, and HBx protein) and four regulatory elements (enhancer II/basal core promoter, preS1 promoter, preS2/S promoter, and enhancer I/X promoter) (Figure 1) [10]. HBx is encoded by ORF-X, which is upstream of ORF-C and near the sticky end of the HBV genome, where it also overlaps with other genes. In HBV genotype B/C, the HBx gene is located at nucleotide (nt.) 1060 to 1838 of the HBV genome. From nt. 1060 to 1373 is the promoter region of HBx, and from nt. 1374 to 1835 is the coding region of HBx [10]. HBx protein consists of 154 amino acids (aa) with a molecular mass of approximately 17kDa, and is commonly situated in the cytoplasm and to a lesser extent in the nucleus of hepatocytes [11]. HBx protein consists of two functional domains. The amino-terminal domain is encoded by the first 50 aa, which can inhibit HBx activities. The trans-activation function domain is located between aa 52-148, of which aa 120-140 is involved in the nuclear trans-activation mechanisms, aa 58-119 is involved in signal transduction activities, and the C-terminal 20 aa is related to the stability of HBx [12].
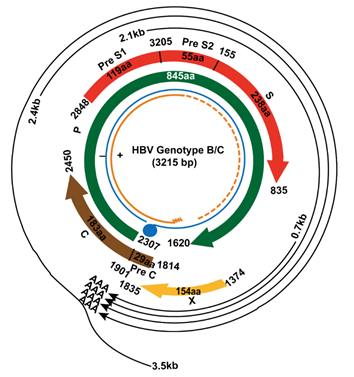
Figure 1: Genome of HBV genotype B/C.
3. HBx Integrated into Host Genome
HBV can integrate into the human genome, thus contributing to genomic instability and hepatocarcinogenesis. Approx-imately 40% of HBV breakpoints in the HBV genome are located within a 1,800-bp region where the viral enhancer, X gene, and core gene are located [13]. HBx integrates into the cancer-related genes such as TERT, MLL4, and CCNE1 and affects their expression [14]. HBV integration can produce HBV-human chimeric transcripts that exert oncogenic effects. HBV-human chimeric transcripts are mainly fusions of HBx gene with repetitive elements within introns of human genes such as long interspersed nuclear elements (LINEs) [15]. HBx-LINE1 chimeric transcript, as a long non-coding RNA, down-regulates the expression of miR-122, leading to increased activity of Wnt/ β-catenin pathway, inducing colony formation, invasion, and migration, and leading to the occurrence and development of tumors [16].
Random HBV genome integration can lead to truncation of the HBV genome, especially at the C-terminal of the HBx gene. The generation of C-terminal truncated HBx (Ct-HBx) is a common event in the occurrence and development of HCC. Many different Ct-HBx have been found in the HBV infector, cirrhosis, and HCC patient, and usually demon-strate a stronger pro-carcinogenic effects compared to the full-length of HBx [17]. Ct-HBx can synergistically down-regulate the expression of TXNIP with NFATC2, leading to glucose metabolism reprogramming, thus initiating the occurrence of HCC and promoting the migration and invasion of cancer cells [18].
4. Mutations of HBx Gene
4.1 APOBEC promotes HBx mutations under inflammation
Chronic inflammation is a prerequisite for the development of cancers. The chronic non-resolving inflammation status of the liver is mainly caused by HBV infection. Under this status, both HBV and host cells undergo an evolutionary process of “mutation–selection–adaptation”, which promo-tes the occurrence and progression of HCC [19, 20]. The family of cytidine deaminases and their analogues called “apolipoprotein B mRNA editing enzyme catalytic polypeptides (APOBECs),” play critical roles in various biologic processes, and are trans-activated by pro-inflammatory molecules [21]. APOBECs can impact HBV replication and induce HBV hypermutation via cytidine deamination [22, 23]. The expression level of APOBEC3s was significantly correlated with HBV quasispecies complexity [24]. Among the HBV genome, APOBEC3s prefer to cause the mutations in HBx [25]. APOBEC3-mediated HBx mutants cause a gain of function that enhances the colony-forming ability and proliferative capacity of neoplastic cells [26].
4.2 Interaction between genetic polymorphisms of inflammatory/immune pathway genes and HBx mutations
Polymorphic genotypes associated with increased risk of chronic progression of HBV infection and significant immune selection of HCC-related HBx mutants were more frequent in Chinese Han populations than in European populations [20]. NF-κB and STAT3 signaling pathways are involved in the occurrence and development of HCC. In the Han population, the variant genotypes of functional single nucleotide polymorphisms (SNPs) rs2233406 and rs3138-053 in the NFKBIA promoter region facilitate the immune selection of the HCC-related HBx mutants including A1762T/G1764A and T1753V, leading to increased risk of HCC. In addition, the interaction of rs2233406 variant genotypes and A1762T/G1764A was significantly associ-ated with increased risk of HCC [27]. STAT3 rs2293152 variant genotype promotes the selection of HBx mutants (A1762T/G1764A and T1674C/G) positively related to the occurrence of HCC and interacts with these mutants to promote HCC development [28]. SNPs in HLA-DP, HLA-DQ, and HLA-DR were associated with chronic HBV infection or response to hepatitis B vaccination in Asians [29, 30]. HLA-DP genotypes (rs3077T, rs3135021A, rs9277535A) that promote HBV clearance are closely related to the HBx cancer-promoting mutants (C1653T and T1674C/G) and are negatively correlated with tumor suppressor HBx mutants (G1652A, T1673C, T1674C, G1719T, G1730C, G1799C, and A1727T) [31]. Our recent study found that HLA-DR variant genotypes of rs3135395, rs477515, and rs3135338 were negatively correlated with the generation of HBx mutants (A1762T/G1764A, T1753V, and C1653T) [32].
4.3 HBx mutants accumulate and show the stronger carcinogenic ability
In our previous research, the wild-type (“standard”) HBV sequences were established, and HCC-related mutations and their development patterns were subsequently identified. The results showed that key HBx mutants, including G1613A, C1653T, T1674C/G, T1753V, and A1762T/ G1764A, gradually accumulate during the development of HCC in hepatitis B patients and are risk factors for HCC development. The combination of HBx mutants can be utilized to predict the occurrence and progression of HCC [33-35]. Mutations in the HBx gene may alter the structure of the HBx protein, thereby affecting oncogenic potential. A1762T/G1764A/T1753A/T1768A mutation can up-regulate Skp2, which then down-regulates P53 via ubiquitin-mediated proteasomal degradation, increasing the risk of hepatocellular carcinoma [36]. In our recent research, we used the Sleeping Beauty (SB) transposon system to deliver HBx wild-type and four HBx mutants (A1762T/G1764A, T1674G/T1753C/A1762T/G1764A, C1653T/T1674G/ A1762T/G1764A, and Ct-HBx) into the livers of fumarylacetoacetate hydrolase (Fah)-deficient mice. In those mouse models, C1653T/T1674G/A1762T/G1764A mutant resulted in a higher HCC incidence and had a stronger capacity of upregulating inflammatory cytokines. C1653T/T1674G/A1762T/G1764A mutant promoted the proliferation of HCC cells by up-regulating PAI1 [37]. Those results indicate that targeting HBx mutations related pathways can help to handle the dilemma of HBV-HCC.
5. HBx and Transcriptional Activity
HBx protein cannot directly bind to the DNA, but can trans-regulate gene transcription via interacting with various protein factors to activate promoters and enhancers, thus affecting the occurrence and development of HCC [12]. ARID2 can inhibit cell cycle progression and tumor growth in HCC, and its promoter region nt-1040/nt-601 contains potential ATOH1 binding elements. HBx inhibited ARID2 expression by impairing binding of the transcription factor ATOH1 to the ARID2 promoter [38]. HBx can stimulate HAT1 promoter by co-activating Sp1 to induce HAT1 expression, contributing to the assembly and epigenetic regulation of HBV cccDNA minichromosomes [39].
6. HBx and Epigenetic Modification
Epigenetic changes affect the expression of coding and non-coding genes, promote HBV replication and HCC develop-ment, and are caused by various factors, such as pathogens, chemicals, and ultraviolet light. HBx is considered to be the most important factors affecting epigenetic inheritance in HBV-HCC. HBx regulates gene expression via regulating its promoter methylation status. HBx upregulates the expres-sion of DNMT3A and interacts with DNMT3A to increase the level of DNA methylation in PTPN13 promoter region and inhibit the transcription of PTPN13 [40]. HBx induces RelA to form complexes with EZH2, TET2, and DNMT3L, resulting in demethylation of CpG site on NF-κB side of EpCAM and up-regulation of EpCAM expression [41. HBx interacts with MBD2 and CBP/p300 to induce the formation of the MBD2-HBx-CBP/p300 complex and mediate the acetylation of histone H3 and H4 [42]. ZHX2 is a tumor suppressor gene associated with HCC, and HBx promotes the expression of miR-155, thereby reducing the level of ZHX2 [43]. Binding of HBx to SKP2 leads to SHIP2 ubiquitination and promotes HCC progression [44].
7. HBx Rolled in the Oncogenic Pathway of HCC
7.1 Wnt/β-catenin pathway
Wnt/β-catenin pathway has important functions in embryo development, and abnormal Wnt signaling can stimulate tumorigenesis. HBx may promote HL-7702 cell proliferation via the COX-2/Wnt/β-catenin pathway [45]. HBx induces miR-5188-FOXO1/β-catenin-C-Jun feedback loop through the Wnt signaling pathway, thus inducing the generation of cancer stem cells [46]. HBx regulates the stem-like properties of OV6+ cancer stem-like cells in HCC via the MDM2/CXCL12/CXCR4/β-catenin signaling axis [47]. HBx mutants, especially the combinatorial mutant, allow constitutive activation of the Wnt signaling pathway and may play a pivotal role in HBV-HCC [48].
7.2 PI3K/AKT pathway
PI3K/AKT pathway is vital in hepatocacinogenesis. HBx activates the autophagic lysosome pathway through the PI3K-Akt-mTOR pathway, and increases the formation of autophagosomes and autolysosomes [49]. HBx induced alpha-fetoprotein (AFP) receptor expressed to activate PI3K/AKT signal to promote expression of Src in liver cells and hepatoma cells [50].
7.3 NF-κB pathway
HBx can facilitate translocation of NF-κB from the cytoplasm to the nucleus, and the binding of NF-κB to the S100A9 promoter enhances the transcription of S100A9. Silencing S100A9 expression partially blocks HBx-induced growth and metastasis of HCC cells [51]. HBx activates NF-κB, which in turn directly drives IFIT3 transcription and enhance HBV replication [52]. HBx promotes IKKβ-induced NF-κB activation by inhibiting miR-34a, and induces phosphorylation of STAT3, which together promote the expression and secretion of HMGB1, and promote EMT progression and angiogenesis in HCC [53].
7.4 Oxidative stress pathway
HBV infection induced endoplasmic reticulum (ER) stress
by chronic inflammation via enhanced inflammation, oxidative stress-mediated DNA damage, and hepatocyte proliferation. Mutation types in four regions of HBV genome (preS1, preS2, S and C) are associated with endoplasmic reticulum stress mechanism [54]. HBx related oxidative stress pathway was also reported in the past few years. In hydrogen peroxide stimulated cells, HBx triggered the release of ASC, IL-1β, IL-18 and initiated pro-inflammatory cell death (pyroptosis). Cells treated with mitoROS scavenger attenuated HBx-induced NLRP3 activation and pyroptosis [55]. It was reported that HBx exerts a pro-apoptotic effect upon exposure to oxidative stress probably by accelerating the loss of Mcl-1 protein via caspase-3 cascade [56]. HBx downregulated the expression of NQO1, thus reducing intracellular glutathione levels, impairing mitochondrial function, and increasing susceptibility of hepatoma cells to oxidative stress-induced cell injury [57].
8. HBx and Metabolism
HBx disrupted the metabolism of glucose, lipids, and amino acids, especially nucleic acids [58]. HBx associates with p62 and the Nrf2 repressor Keap1 to form HBx-p62-Keap1 complex in the cytoplasm. The aggregation of HBx-p62-Keap1 complexes hijack Keap1 from Nrf2, leading to the activation of Nrf2 and consequently G6PD transcription [59]. HBx induced BNIP3L-dependent mitophagy which upregulated glycolytic metabolism, increasing cancer stemness of HCC cells in vivo and in vitro [60]. HBx up-regulates SWELL1 through co-activating transcription factor Sp1, regulating arachidonic acid metabolism signaling [61]. HBx and COXIII co-localization in HL-7702 cells lead to upregulation of the mitochondrial function and ROS generation [62].
9. HBx and Immune Tolerance
During chronic HBV infection in humans, the adaptive immunity changes from immune tolerance to progressive immune activation, inactivation, reactivation, and exhau-stion, all of which may be the immune pathogenic factors for the development of HCC [63]. Complex interplay between HBx-deregulated miRNAs and immune responses affects HBV-HCC development [5]. Death receptors of TNFSF10/ TRAIL contribute to immune surveillance against virus-infected or transformed cells by promoting apoptosis. HBx restricts TNFSF10 receptor signaling via macroauto-phagy/autophagy-mediated degradation of TNFRSF10B/ DR5, thereby enabling HBV to evade antiviral immunity [64]. HBx promotes HBV immune escape by inhibiting transcription of TRIM22 through methylating its 5'-UTR and inhibiting IFN-stimulated TRIM22 [65].
10. HBx and HCC Prevention and Treatment
High viral load is associated with poor postoperative prognosis of HBV-HCC, and antiviral therapy can reduce HCC recurrence and related deaths significantly. Antiviral therapy can also modulate hepatocarcinogenesis by decree-sing the levels of HBx to inhibit the tumorigenic effect of MSL2 and cccDNA [66]. However, the effect of antiviral therapy is not significant for patients with Ct-HBx [67]. During long-term treatment, the efficacy of nucleoside analogues is diminished by the presence of resistant mutants. HBx mutations in drug-resistant patients lead to increased cccDNA levels to compensate for replication suppression [68]. HBx is necessary for HBV replication, hence HBV replication can be inhibited by reducing the expression of HBx. Dicoumarol, an inhibitor of NAD(P)H quinone oxidoreductase 1 (NQO1), significantly reduced HBx expression thus has a role in HBV replication [69]. HBx-based vaccines eliminate persistent HBV in animal models and have the potential to be developed as a therapeutic vaccine against chronic hepatitis B [70].
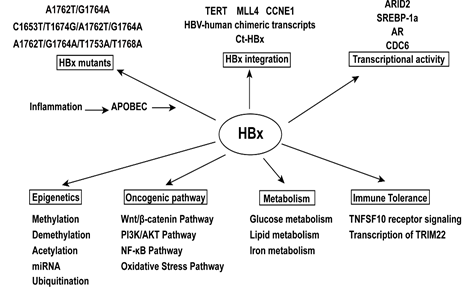
Figure 2: The mechanism of hepatitis B virus X gene in promoting hepatocellular carcinoma.
11. Conclusion
HBx gene and its encoded protein are widely involved in the process of chronic hepatitis, cirrhosis, and liver cancer by promoting HBV replication, integration into the host genome, and evolution. The inflammation microenviron-ment interacting with APOBECs can both promote HBV and host cells evolution, which leads to poor prognosis of HCC patients. Epidemiological evidence indicates that HBx mutants are closely related to HBV-HCC and gradually accumulate during the development of HCC. Mutant HBx has a stronger carcinogenic ability than wild-type HBx and is associated with antiviral treatment resistance. In recent years, HBx has been found to play an important role in the occurrence, recurrence, metastasis and immune escape of HCC via numerous new mechanisms, such as trans-activation and trans-repression, epigenetic modification, activation of oncogenic pathways, metabolic disorders and drug resistance. However, the specific mechanism of mutant HBx in the occurrence, development, and drug resistance of HCC is still less studied. The knowledge of details of HBx related evolution is far from satisfactory. In the future, further research on the mechanism of HBx and its mutant is needed, and the development of drugs targeting HBx mutant will be of great significance for the prevention, treatment, and prognosis prediction of HBV-HCC.
Acknowledgements
Not applicable.
Funding
This work was supported by grant 2015CB554006 from the National Key Basic Research Program of China (GC); grants 91529305 (GC), 81520108021 (GC), 81673250 (GC) from the National Natural Science Foundation of China.
Conflict of Interest
The authors declare that they have no competing interests.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 71 (2021): 209-249.
- Wang S, Du X, Han X, et al. Influence of socioeconomic events on cause-specific mortality in urban Shanghai, China, from 1974 to 2015: a population-based longitudinal study. CMAJ 190 (2018): E1153-E1161.
- Jiang D, Zhang L, Liu W, et al. Trends in cancer mortality in China from 2004 to 2018: A nationwide longitudinal study. Cancer Commun (Lond) 41 (2021): 1024-1036.
- Yang F, Ma L, Yang Y, et al. Contribution of Hepatitis B Virus Infection to the Aggressiveness of Primary Liver Cancer: A Clinical Epidemiological Study in Eastern China. Front Oncol 9 (2019): 370.
- Sartorius K, An P, Winkler C, et al. The Epigenetic Modulation of Cancer and Immune Pathways in Hepatitis B Virus-Associated Hepatocellular Carcinoma: The Influence of HBx and miRNA Dysregulation. Front Immunol 12 (2021): 661204.
- Slagle BL, Bouchard MJ. Role of HBx in hepatitis B virus persistence and its therapeutic implications. Curr Opin Virol 30 (2018): 32-38.
- Chauhan R, Michalak TI. Earliest hepatitis B virus-hepatocyte genome integration: sites, mechanism, and significance in carcinogenesis. Hepatoma Research 7 (2021): 20.
- Ali A, Abdel-Hafiz H, Suhail M, et al. Hepatitis B virus, HBx mutants and their role in hepatocellular carcinoma. World J Gastroenterol 20 (2014): 10238-10248.
- Geng M, Xin X, Bi LQ, et al. Molecular mechanism of hepatitis B virus X protein function in hepatocarcinogenesis. World J Gastroenterol 21 (2015): 10732-10738.
- Jiang Y, Han Q, Zhao H, et al. The Mechanisms of HBV-Induced Hepatocellular Carcinoma. J Hepatocell Carcinoma 8 (2021): 435-450.
- Mathew MA, Kurian SC, Varghese AP, et al. HBx Gene Mutations in Hepatitis B Virus and Hepatocellular Carcinoma. Gastroenterology Res 7 (2014): 1-4.
- Slagle BL, Bouchard MJ. Hepatitis B Virus X and Regulation of Viral Gene Expression. Cold Spring Harb Perspect Med 6 (2016): a021402.
- Sung WK, Zheng H, Li S, et al. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat Genet 44 (2012): 765-769.
- Zhao LH, Liu X, Yan HX, et al. Genomic and oncogenic preference of HBV integration in hepatocellular carcinoma. Nat Commun 7 (2016): 12992.
- Jin Y, Lee WY, Toh ST, et al. Comprehensive analysis of transcriptome profiles in hepatocellular carcinoma. J Transl Med 17 (2019): 273.
- Liang HW, Wang N, Wang Y, et al. Hepatitis B virus-human chimeric transcript HBx-LINE1 promotes hepatic injury via sequestering cellular microRNA-122. J Hepatol 64 (2016): 278-291.
- Sze KM, Chu GK, Lee JM, et al. C-terminal truncated hepatitis B virus x protein is associated with metastasis and enhances invasiveness by C-Jun/matrix metalloproteinase protein 10 activation in hepatocellular carcinoma. Hepatology 57 (2013): 131-139.
- Zhang Y, Yan Q, Gong L, et al. C-terminal truncated HBx initiates hepatocarcinogenesis by downregulating TXNIP and reprogramming glucose metabolism. Oncogene 40 (2021): 1147-1161.
- Liu WB, Wu JF, Du Y, et al. Cancer Evolution-Development: experience of hepatitis B virus-induced hepatocarcinogenesis. Curr Oncol 23 (2016): e49-e56.
- Cao G-W. Cancer Evo-Dev, a novel hypothesis derived from studies on hepatitis B virus-induced carcinogenesis. Hepatoma Research 3 (2017).
- Gao J, Choudhry H, Cao W. Apolipoprotein B mRNA editing enzyme catalytic polypeptide-like family genes activation and regulation during tumorigenesis. Cancer Sci 109 (2018): 2375-2382.
- Zhang Y, Chen X, Cao Y, et al. Roles of APOBEC3 in hepatitis B virus (HBV) infection and hepatocarcinogenesis. Bioengineered 12 (2021): 2074-2086.
- Deng Y, Du Y, Zhang Q, et al. Human cytidine deaminases facilitate hepatitis B virus evolution and link inflammation and hepatocellular carcinoma. Cancer Lett 343 (2014): 161-171.
- Yin J, Chen X, Li N, et al. Compartmentalized evolution of hepatitis B virus contributes differently to the prognosis of hepatocellular carcinoma. Carcinogenesis 42 (2021): 461-470.
- Beggel B, Münk C, Däumer M, et al. Full genome ultra-deep pyrosequencing associates G-to-A hypermutation of the hepatitis B virus genome with the natural progression of hepatitis B. J Viral Hepat 20 (2013): 882-889.
- Xu R, Zhang X, Zhang W, et al. Association of human APOBEC3 cytidine deaminases with the generation of hepatitis virus B x antigen mutants and hepatocellular carcinoma. Hepatology 46 (2007): 1810-1820.
- Zhang Q, Ji XW, Hou XM, et al. Effect of functional nuclear factor-kappaB genetic polymorphisms on hepatitis B virus persistence and their interactions with viral mutations on the risk of hepatocellular carcinoma. Ann Oncol 25 (2014): 2413-2419.
- Xie J, Zhang Y, Zhang Q, et al. Interaction of signal transducer and activator of transcription 3 polymorphisms with hepatitis B virus mutations in hepatocellular carcinoma. Hepatology (Baltimore, Md) 57 (2013): 2369-2377.
- Ji X, Zhang Q, Li B, et al. Impacts of human leukocyte antigen DQ genetic polymorphisms and their interactions with hepatitis B virus mutations on the risks of viral persistence, liver cirrhosis, and hepatocellular carcinoma. Infect Genet Evol 28 (2014): 201-209.
- Chen Y, Lin J, Deng Y, et al. Association of human leukocyte antigen-DR-DQ-DP haplotypes with the risk of hepatitis B virus-related hepatocellular carcinoma. Hepatoma Research 8 (2022): 8.
- Zhang Q, Yin J, Zhang Y, et al. HLA-DP polymorphisms affect the outcomes of chronic hepatitis B virus infections, possibly through interacting with viral mutations. Journal of virology 87 (2013): 12176-12186.
- Deng Y, Li P, Liu W, et al. The genetic polymorphism down-regulating HLA-DRB1 enhancer activity facilitates HBV persistence, evolution and hepatocarcinogenesis in the Chinese Han population. J Viral Hepat 27 (2020): 1150-1161.
- Yin J, Xie J, Liu S, et al. Association between the various mutations in viral core promoter region to different stages of hepatitis B, ranging of asymptomatic carrier state to hepatocellular carcinoma. Am J Gastroenterol 106 (2011): 81-92.
- Yin J, Xie J, Zhang H, et al. Significant association of different preS mutations with hepatitis B-related cirrhosis or hepatocellular carcinoma. J Gastroenterol 45 (2010): 1063-1071.
- Liu S, Xie J, Yin J, et al. A matched case-control study of hepatitis B virus mutations in the preS and core promoter regions associated independently with hepatocellular carcinoma. J Med Virol 83 (2011): 45-53.
- Yan J, Yao Z, Hu K, et al. Hepatitis B Virus Core Promoter A1762T/G1764A (TA)/T1753A/T1768A Mutations Contribute to Hepatocarcinogenesis by Deregulating Skp2 and P53. Dig Dis Sci 60 (2015): 1315-1324.
- Pu R, Liu W, Zhou X, et al. The Effects and Underlying Mechanisms of Hepatitis B Virus X Gene Mutants on the Development of Hepatocellular Carcinoma. Frontiers in Oncology (2022).
- Gao Q, Wang K, Chen K, et al. HBx protein-mediated ATOH1 downregulation suppresses ARID2 expression and promotes hepatocellular carcinoma. Cancer Sci 108 (2017): 1328-1337.
- Yang G, Feng J, Liu Y, et al. HAT1 signaling confers to assembly and epigenetic regulation of HBV cccDNA minichromosome. Theranostics 9 (2019): 7345-7358.
- Yan Y, Huang P, Mao K, et al. Anti-oncogene PTPN13 inactivation by hepatitis B virus X protein counteracts IGF2BP1 to promote hepatocellular carcinoma progression. Oncogene 40 (2021): 28-45.
- Fan H, Zhang H, Pascuzzi PE, et al. Hepatitis B virus X protein induces EpCAM expression via active DNA demethylation directed by RelA in complex with EZH2 and TET2. Oncogene 35 (2016): 715-726.
- Liu XY, Tang SH, Wu SL, et al. Epigenetic modulation of insulin-like growth factor-II overexpression by hepatitis B virus X protein in hepatocellular carcinoma. Am J Cancer Res 5 (2015): 956-978.
- Song X, Tan S, Wu Z, et al. HBV suppresses ZHX2 expression to promote proliferation of HCC through miR-155 activation. Int J Cancer 143 (2018): 3120-3130.
- Su K-J, Yu Y-L. Downregulation of SHIP2 by Hepatitis B Virus X Promotes the Metastasis and Chemoresistance of Hepatocellular Carcinoma through SKP2. Cancers (Basel) 11 (2019).
- Zheng BY, Gao WY, Huang XY, et al. HBx promotes the proliferative ability of HL7702 cells via the COX2/Wnt/betacatenin pathway. Mol Med Rep 17 (2018): 8432-8438.
- Lin X, Zuo S, Luo R, et al. HBX-induced miR-5188 impairs FOXO1 to stimulate β-catenin nuclear translocation and promotes tumor stemness in hepatocellular carcinoma. Theranostics 9 (2019): 7583-7598.
- Wang C, Wang MD, Cheng P, et al. Hepatitis B virus X protein promotes the stem-like properties of OV6(+) cancer cells in hepatocellular carcinoma. Cell Death Dis 8 (2017): e2560.
- Chen Z, Tang J, Cai X, et al. HBx mutations promote hepatoma cell migration through the Wnt/beta-catenin signaling pathway. Cancer Sci 107 (2016): 1380-1389.
- Wang P, Guo QS, Wang ZW, et al. HBx induces HepG-2 cells autophagy through PI3K/Akt-mTOR pathway. Mol Cell Biochem 372 (2013): 161-168.
- Zhu M, Guo J, Li W, et al. HBx induced AFP receptor expressed to activate PI3K/AKT signal to promote expression of Src in liver cells and hepatoma cells. BMC Cancer 15 (2015): 362.
- Duan L, Wu R, Zhang X, et al. HBx-induced S100A9 in NF-kappaB dependent manner promotes growth and metastasis of hepatocellular carcinoma cells. Cell Death Dis 9 (2018): 629.
- Xu F, Song H, An B, et al. NF-kappaB-Dependent IFIT3 Induction by HBx Promotes Hepatitis B Virus Replication. Front Microbiol 10 (2019): 2382.
- Zhang Y, Ren H, Li J, et al. Elevated HMGB1 expression induced by hepatitis B virus X protein promotes epithelial-mesenchymal transition and angiogenesis through STAT3/miR-34a/NF-κB in primary liver cancer. Am J Cancer Res 11 (2021): 479-494.
- Choi YM, Lee SY, Kim BJ. Naturally Occurring Hepatitis B Virus Mutations Leading to Endoplasmic Reticulum Stress and Their Contribution to the Progression of Hepatocellular Carcinoma. Int J Mol Sci 20 (2019).
- Xie WH, Ding J, Xie XX, et al.Hepatitis B virus X protein promotes liver cell pyroptosis under oxidative stress through NLRP3 inflammasome activation. Inflamm Res 69 (2020): 683-696.
- Hu L, Chen L, Yang G, et al. HBx sensitizes cells to oxidative stress-induced apoptosis by accelerating the loss of Mcl-1 protein via caspase-3 cascade. Mol Cancer 10 (2011): 43.
- Wu YL, Wang D, Peng XE, et al. Epigenetic silencing of NAD(P)H:quinone oxidoreductase 1 by hepatitis B virus X protein increases mitochondrial injury and cellular susceptibility to oxidative stress in hepatoma cells. Free Radic Biol Med 65 (2013): 632-644.
- Dan Y, Zhang Y, Cheng L, et al. Hepatitis B virus X protein (HBx)-induced abnormalities of nucleic acid metabolism revealed by (1)H-NMR-based metabonomics. Sci Rep 6 (2016): 24430.
- Liu B, Fang M, He Z, et al. Hepatitis B virus stimulates G6PD expression through HBx-mediated Nrf2 activation. Cell Death Dis 6 (2015): e1980.
- Chen Y-Y, Wang W-H, Che L, et al. BNIP3L-Dependent Mitophagy Promotes HBx-Induced Cancer Stemness of Hepatocellular Carcinoma Cells via Glycolysis Metabolism Reprogramming. Cancers (Basel) 12 (2020).
- Liu Z, Wang J, Liu L, et al. Chronic ethanol consumption and HBV induce abnormal lipid metabolism through HBx/SWELL1/arachidonic acid signaling and activate Tregs in HBV-Tg mice. Theranostics 10 (2020): 9249-9267.
- Zou LY, Zheng BY, Fang XF, et al. HBx co-localizes with COXIII in HL-7702 cells to upregulate mitochondrial function and ROS generation. Oncol Rep 33 (2015): 2461-2467.
- Chen Y, Tian Z. HBV-Induced Immune Imbalance in the Development of HCC. Front Immunol 10 (2019): 2048.
- Shin G-C, Kang HS, Lee AR, et al. Hepatitis B virus-triggered autophagy targets TNFRSF10B/death receptor 5 for degradation to limit TNFSF10/TRAIL response. Autophagy 12 (2016): 2451-2466.
- Lim K-H, Park E-S, Kim DH, et al. Suppression of interferon-mediated anti-HBV response by single CpG methylation in the 5'-UTR of. Gut 67 (2018): 166-178.
- Jin XL, Hong SK, Kim H, et al. Antiviral therapy may decrease HBx, affecting cccDNA and MSL2 in hepatocarcinogenesis. Oncol Lett 18 (5): 4984-4991.
- Yin J, Li N, Han Y, et al. Effect of antiviral treatment with nucleotide/nucleoside analogs on postoperative prognosis of hepatitis B virus-related hepatocellular carcinoma: a two-stage longitudinal clinical study. J Clin Oncol 31(2013): 3647-3655.
- Lin C-L, Chien R-N, Chu Y-D, et al. Hepatitis B virus X gene mutants emerge during antiviral therapy and increase cccDNA levels to compensate for replication suppression. Hepatol Int 14 (2020): 973-984.
- Cheng ST, Hu JL, Ren JH, et al. Dicoumarol, an NQO1 inhibitor, blocks cccDNA transcription by promoting degradation of HBx. J Hepatol 74 (2021): 522-534.
- Horng J-H, Lin W-H, Wu C-R, et al. HBV X protein-based therapeutic vaccine accelerates viral antigen clearance by mobilizing monocyte infiltration into the liver in HBV carrier mice. J Biomed Sci 27 (2020): 70.


 Impact Factor: * 4.1
Impact Factor: * 4.1 Acceptance Rate: 74.74%
Acceptance Rate: 74.74%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 