Molecular Pathogen Testing for Identifying the Etiology of Febrile Illness in Immunocompromised Children
Fabian JS van der Velden1,2, Priyen Shah3, Marie Voice4, Emma Lim1, Jethro Herberg3, Victoria Wright3, Tisham De3, Enitan D. Carrol5,6, Aakash Khanijau5, Andrew J Pollard7, Stéphane Paulus7, Ulrich von Both8, Laura Kolberg8, Clementien L Vermont9, Nienke N Hagedoorn9, Federico Martinón-Torres10,11,12, Irene Rivero Calle10, Philipp KA Agyeman13, Luregn J Schlapbach14, Maria Tsolia15, Irini Eleftheriou15, Marko Pokorn16, Mojca Kolnik16, Taco W Kuijpers17, Dace Zavadska18, Aleksandra Rudzate18, Nina A Schweintzger19, Werner Zenz19, Shunmay Yeung20, Michiel van der Flier21,22, Ronald de Groot21, Michael Levin3, Colin Fink4, Marieke Emonts1,2,23,*, on behalf of the PERFORM consortium**
1Great North Children’s Hospital, Paediatric Immunology, Infectious Diseases & Allergy, The Newcastle upon Tyne Hospitals NHS Foundation Trust, Newcastle upon Tyne, United Kingdom
2Translational and Clinical Research Institute, Newcastle University, Newcastle upon Tyne, United Kingdom
3Wright-Fleming Institute, Section of Paediatric Infectious Disease, Imperial College London, London, United Kingdom
4Micropathology Ltd, University of Warwick, Warwick, United Kingdom
5Institute of Infection, Veterinary and Ecological Sciences, University of Liverpool, Liverpool, United Kingdom
6Alder Hey Children’s NHS Foundation Trust, Liverpool, United Kingdom
7Oxford Vaccine Group, Department of Paediatrics, University of Oxford, Oxford, United Kingdom
8Division Paediatric Infectious Diseases, Dr. Von Hauner Children’s Hospital, University Hospital, LMU, Munich, Germany
9Department of Pediatrics, Division of Pediatric Infectious Diseases & Immunology, Erasmus MC-Sophia Children’s Hospital, Rotterdam, The Netherlands
10Pediatrics Department, Translational Pediatrics and Infectious Diseases, Hospital Clínico Universitario de Santiago, Santiago de Compostela, Spain
11GENVIP Research Group, Instituto de Investigación Sanitaria de Santiago, Universidad de Santiago de Compostela, Santiago de Compostela, Spain
12Consorcio Centro de Investigacion Biomedicaen Red de Enfermedades Respiratorias (CIBERES), Madrid, Spain
13Department of Pediatrics, Inselspital, Bern University Hospital, University of Bern, Bern, Switzerland
14 Department of Intensive Care and Neonatology, and Children’s Research Center, University Children’s Hospital Zürich, Zürich, Switzerland
152nd Department of Pediatrics, National and Kapodistrian University of Athens, Children’s Hospital ‘P, and A. Kyriakou’, Athens, Greece
16University Children’s Hospital, University Medical Centre Ljubljana, Ljubljana, Slovenia
17Amsterdam University Medical Center, location Academic medical Center, Department of Pediatric Immunology, Rheumatology and Infectious Diseases, University of Amsterdam, Amsterdam, The Netherlands
18Department of Pediatrics, R?gas Stradi?a Universit?te, Children’s Clinical University Hospital, Riga, Latvia
19Department of Pediatrics and Adolescent Medicine, Division of General Pediatrics, Medical University of Graz, Graz, Austria
20Clinical Research Department, Faculty of Infectious and Tropical Disease, London School of Hygiene and Tropical Medicine, London, United Kingdom
21Pediatric Infectious Diseases and Immunology, Amalia Children’s Hospital, Radboud University Medical Center, Nijmegen, The Netherlands
22Pediatric Infectious Diseases and Immunology, Wilhelmina Children’s Hospital, University Medical Center Utrecht, Utrecht, The Netherlands
23NIHR Newcastle Biomedical Research Centre, Newcastle upon Tyne Hospitals NHS Foundation Trust and Newcastle University, Newcastle upon Tyne, United Kingdom
*Corresponding author: Marieke Emonts, Great North Children’s Hospital, Paediatric Immunology, Infectious Diseases and Allergy, The Newcastle upon Tyne Hospitals NHS Foundation Trust, Newcastle upon Tyne, United Kingdom.
**Members of the PERFORM Consortium are listed in the supplementary files
Received: 01 August 2023; Accepted: 09 August 2023; Published: 14 September 2023
Article Information
Citation: Fabian JS van der Velden, Priyen Shah, Marie Voice, Emma Lim, Jethro Herberg, Victoria Wright, Tisham De, Enitan D. Carrol, Aakash Khanijau, Andrew J Pollard, Stéphane Paulus, Ulrich von Both, Laura Kolberg, Clementien L Vermont, Nienke N Hagedoorn, Federico Martinón- Torres, Irene Rivero Calle, Philipp KA Agyeman, Luregn J Schlapbach, Maria Tsolia, Irini Eleftheriou, Marko Pokorn, Mojca Kolnik, Taco W Kuijpers, Dace Zavadska, Aleksandra Rudzate, Nina A Schweintzger, Werner Zenz, Shunmay Yeung, Michiel van der Flier, Ronald de Groot, Michael Levin, Colin Fink, Marieke Emonts, on behalf of the PERFORM consortium. Molecular Pathogen Testing for Identifying the Etiology of Febrile Illness in Immunocompromised Children. Journal of Biotechnology and Biomedicine. 6 (2023): 380-391.
DOI: 10.26502/jbb.2642-91280100
View / Download Pdf Share at FacebookAbstract
Background: Diagnosing febrile illness in immunocompromised children at presentation to hospital remains a challenge. Serious bacterial infection can cause significant mortality and morbidity, but conventional diagnostics using culture-based technology results are often negative. Molecular pathogen testing might increase the yield of pathogen detection in this population, subsequently potentially altering clinical management and improving outcome.
Methods: Immunocompromised febrile children recruited to the international Personalised Risk assessment in Febrile illness to Optimised Real-life Management study (PERFORM) were evaluated using current best practice local diagnostic approaches, and subsequently assigned to phenotypes based on standardized definitions. Retrospectively, these febrile cases were complemented by additional centralized molecular tests (CMT) for 22 respiratory and 35 blood pathogens and subsequently we analyzed febrile cases using CMT and local microbiological data. Results: There were 336 febrile episodes, of which 45 definite bacterial (13.4%), 37 definite viral (11.0%) using conventional diagnostic approaches, and 254 with more uncertain or inflammatory etiologies of fever (75.6%), and 54 non-febrile control cases. CMT detected any pathogens in 201/336 febrile cases (59.8%) and 33/54 non-febrile controls (61.1%). CMT detected E. cloacae, Enterobacteriales, and K. pneumoniae more commonly in definite bacterial cases compared to other febrile cases. There were no pathogens significantly more often detected in febrile cases versus controls, nor were any viral pathogens more often detected in definite viral cases by CMT. Bacterial pathogen detection was seen more often in definite bacterial cases on CMT (odds ratio 20.71 (95% confidence interval 2.11-203.77)). Viruses were detected in 46.7% of definite bacterial cases (N= 21), and 57.4% of controls (N= 31). Bacteria were detected in 10.8% of definite viral cases (N= 4), and 5.6% of controls (N=3). Human herpes virus (HHV)7 was equally the most commonly detected pathogen across all phenotypes (33.6%, N=113) in respectively 22.2% of definite bacterial (N=10), 32.4% of definite viral (N=12), and 36.4% of unknown bacterial or viral febrile illness (N=43) and controls (37.0%, N=20).
Conclusion: CMT frequently detected pathogens in both febrile children and non-febrile controls. Except for certain gram-negative bacteria, no pathogens were more commonly detected in definite bacterial or definite viral febrile illness. Viruses are detected in a significant proportion of patients with bacterial infection. HHV7 is frequently detected in immunocompromised children, both febrile and non-febrile. CMT can increase detection of pathogens, but our data do not suggest it will ease the current diagnostic challenges regarding their clinical relevance in this population.
Keywords
<p>Immunocompromised; Children; Pediatrics; Fever; Infection; Diagnostics</p>
Article Details
Abbreviations:
BIVA-HR: Biomarker Validation in high-risk patients; CI: Confidence Interval; CMT: Centralized molecular tests; EBV: Epstein-Barr virus; ED: Emergency department; HHV: Human herpes virus; OR: Odds ratio; PCR: Polymerase chain reaction; PERFORM: Personalised Risk assessment in Febrile illness to Optimised Real-life Management study; SBI: Serious bacterial infection
Introduction
Febrile illness is a common problem in the pediatric emergency department (ED) [1]. Children with complex comorbidities are frequently seen and are at increased risk of serious infections, for example, due to being immunocompromised or being dependent on central venous lines [2]. It has been estimated that febrile illness accounts for up to 60% of ED attendances in pediatric cancer [3]. These children are at high-risk of serious bacterial infection (SBI) and life-threatening infectious complications [2,4]. Some of these patients are neutropenic. SBI during febrile neutropenia is associated with significant morbidity and mortality if left untreated and therefore it is managed as a medical emergency [2,5]. Due to this significant risk, immunocompromised children with febrile illness are virtually always admitted for broad-spectrum antibiotic treatment, whilst awaiting microbiological results [6,7]. This approach has significantly reduced morbidity and mortality [8], but consequently leads to antibiotic overuse, prolonged hospitalization and increased risk of antimicrobial resistance. In only 11-31% of these patients a microbiologically documented infection will be identified [9-13]. and up to 62% will have no cause identified [11,14-16]. It is estimated that a significant proportion of fever in this population is due to self-limiting viral infection, drug-induced or caused by the underlying condition [5,17,18]. This diagnostic uncertainty incurs significant costs to healthcare systems [19]. Differentiating between bacterial, viral, inflammatory or other causes of fever is challenging, as the clinical presentation is often non-specific [20]. Microbiological cultures take at least 24-48 hours to yield results, and commonly used inflammatory markers such as C-reactive protein and procalcitonin are not sensitive enough to rule out bacterial infection [21-23]. Thus, new approaches are needed to earlier differentiate the etiology of fever, or facilitate early discontinuation of antibiotic treatment. Many biomarkers have been studied, but these have not proven to be sensitive or specific enough to change current practice [24,25]. As an alternative approach, molecular methods can rapidly detect RNA or DNA of bacterial, viral and fungal pathogens [26-29], and might have the potential to change the diagnosis and management of febrile illness. In patients with febrile neutropenia [30-33], immunocompetent children [34,35] or suspected sepsis [26,36] potential benefit of polymerase chain reaction (PCR) has been described, improving the detection rate of pathogens. This study aimed to explore the potential role of centralized molecular pathogen tests (CMT) by PCR in children at high risk of SBI, recruited to the Biomarker Validation in high-risk patients (BIVA-HR) cohort of the Personalized Risk assessment in Febrile illness to Optimise Real-life Management (PERFORM) study (www.perform2020.org), evaluating the diagnosis of fever in children across Europe using current local best practice diagnostics.
Material and Methods
We analyzed data from the BIVA-HR cohort within the PERFORM study [13]. The study recruited patients between 2 June 2016 and 31 December 2019. Children were recruited to BIVA-HR if they fulfilled the following inclusion criteria: <18 years of age, immunocompromised due to primary or secondary immunodeficiency with a (history of) fever (<72 hours prior to admission, T≥38.0°C) or suspected infection, and clinical indication for blood investigations as per treating clinician’s decision. For control patients, similar inclusion criteria were used, with the exception that a (history of) fever or a vaccination <3 weeks from research sample collection were now used as exclusion criteria. These control patients presented to hospital for any reason other than febrile illness, but had a requirement for blood investigations. They received routine clinical workup, according to best practice at the respective centers. Imaging, laboratory and (conventional) microbiological investigations were performed as clinically indicated. For each child a final phenotype was assigned using the validated algorithm in the PERFORM protocol (Supplementary Figure 1), previously described by Nijman et al. [37,38], by experienced pediatricians based on all clinical, laboratory and imaging data in each center. We combined febrile episodes with the phenotypes definite bacterial, probable bacterial, and bacterial syndrome to proven/presumed viral for additional assessment. This was done to reflect the clinical presentation more truthfully at presentation, and to include episodes in which there was a bacterial infection from non-sterile sites or bacterial infections in which inflammatory markers were low. Similarly, we combined febrile episodes with the phenotypes definite viral, probable viral and viral syndrome to proven/presumed viral infection. For similar reasons as in bacterial, this is to be able to include viral infections with higher inflammatory markers, and to reflect the clinical spectrum of viral illness as seen on admission.
Centralized Molecular pathogen Tests (CMT)
Research blood and respiratory samples were obtained from participants as early as possible in the patient’s presentation to hospital, when feasible, simultaneously with the first clinically indicated blood investigations. Collected blood in EDTA and dry flocked throat swabs stored in eNAT® were stored at -80°C and transported to Micropathology Ltd, Coventry, UK. Centralized Molecular pathogen tests were performed retrospectively, in a research setting, and these results were not used for clinical management, local diagnostics, nor final phenotype assignment. Total nucleic acid was extracted from the throat swab and blood samples. Throat swabs were analyzed by NxTAG™ Respiratory Pathogen Panel (RPP) assay (Luminex® Corporation), allowing simultaneous detection of nineteen viral and three bacterial species. Whole blood samples were analyzed utilizing a panel of fourteen multiplex probe-based qPCR assays (Micropathology Ltd, Coventry), containing 23 bacterial, nine viral, and three fungal targets (Supplementary Table 1). Targets were specifically tailored pathogens known to be common in the immunocompromised population. Results from CMT are reported separately from local microbiological investigations, which included molecular testing for common pathogens for some patients.
Table 1: Clinical characteristics of febrile episodes by phenotype, and controls. Data presented as N= (%) or median (interquartile range).
Analysis and statistics
Clinical and laboratory data were collected and final phenotypes were assigned by the local teams on standardized forms and put on a custom online database without personal identifiers. Data quality control and crosschecking for inconsistencies were regularly performed. Descriptive data were reported using absolute frequencies and percentages. Data were not normally distributed; hence, non-parametric tests, medians and interquartile ranges (IQR) were used. Mann-Whitney U tests for continuous variables, and χ2 or Fisher’s exact tests for categorical data, were utilized appropriately. Odds ratios (ORs) were calculated using univariate logistic regression and reported with 95% confidence intervals (CIs). P-values <0.05 were considered statistically significant. Data were analyzed using IBM SPSS Statistics, version 27 [Armonk USA 2020].
Results
During the study period, 599 febrile episodes were enrolled in BIVA-HR [13]. After the removal of episodes without available research samples, 336 febrile episodes were included in the analysis, as well as 54 HR controls. All febrile episodes had blood and respiratory samples, 16 controls had both samples, and 38 controls blood samples only. Following the analysis of clinical and local investigation data, all episodes were assigned a final phenotype. 45 were classified as definite bacterial (13.4%) and 37 as definite viral (11.0%). A further 32 were classified as probable bacterial (9.5%), 19 as bacterial syndrome (5.7%), and 23 as probable viral (6.8%) and 12 as viral syndrome (3.6%). Combining these numbers, 96 episodes (28.6%) were proven/presumed bacterial and 72 episodes (21.4%) were proven/presumed viral. The remaining episodes had other less certain infectious or inflammatory phenotypes assigned (Figure 1). The clinical details of the patients are shown in (Table 1). The most common underlying condition was hematological malignancy (N=140, 41.7%). 131 (38.9%) patients were neutropenic at admission, and 37 (11%) had undergone hematopoietic stem cell transplantation. A high proportion of children were empirically treated with antibiotics at admission, including those with a viral diagnosis (100% of definite bacterial (N=45), 83.7% definite viral (N=31)).
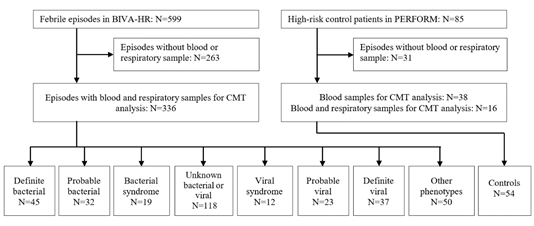
Figure 1: Patient flow and numbers from recruitment to final phenotype assignment based on local investigations and microbiological testing.
CMT pathogen detection in febrile episodes and controls
In 189/336 febrile episodes (56.3%) CMT detected at least one pathogen and 94 (28.0%) had multiple pathogens detected (Table 2). For controls, this was 61.1% (N=31) and 24.1% (N=13) respectively. Human herpes virus (HHV)7 was most commonly detected in both groups, 33.6% of febrile episodes (N=113) and 37.0% of controls (N=20), followed by HHV6b in 12.5% of febrile episodes (N=42) and 24.1% of controls (N=13), and, Epstein-Barr Virus (EBV) in 11.0% (N=37) and 11.1% (N=6) respectively. No statistical differences were observed between cases and controls (Figure 2A), other than detection of HHV6b being more common in controls than in cases (OR 0.45 (IQR 0.22-0.91). The whole spectrum of pathogens detected by local investigations and CMT can be found in Supplementary Tables 2 and 3, respectively.
Table 2: Detection rates of pathogens by CMT in groups, stratified by phenotype. Data presented as N= (%).
CMT pathogen detection in definite bacterial and viral groups, and proven/presumed bacterial and viral groups
Detection of any bacteria with CMT was more commonly observed in the definite bacterial group OR 4.13 (95%CI 1.23-13.82) compared to the definite viral group. Viruses were detected by CMT in 21 definite bacterial episodes (46.7%) and 24 definite viral episodes (64.9%). Vice versa bacteria were detected in 4 definite viral cases (10.8%) and 15 definite bacterial cases (33.3%). Virus detection was not signficantly more common in the definite viral group OR 0.47 (95%CI 0.19-1.16) versus definite bacterial (Figure 2B). When comparing proven/presumed bacterial with proven/presumed viral phenotypes, only detection of blood bacteria was more commonly seen in proven/presumed bacterial infections (OR 2.60 (95%CI 1.04-6.51)) (Figure 2C). Assessing CMT detection in more detail, in 13 definite viral and 4 definite bacterial cases, respiratory viruses were detected by CMT. Most common and the only virus detected in both groups was rhinovirus (detected 3 times each), but it was not significantly more common in either group. No respiratory bacteria were detected in definite bacterial or definite viral cases. Given low detection numbers for the other targets across the groups, no meaningful further comparisons could be made. For blood viruses, 20 definite viral (54.1%) and 19 definite bacterial cases (42.2%) had blood viruses detected. In both groups HHV7 (OR 0.60 (95%CI 0.22-1.59)) and HHV6b (OR 1.52 (95%CI 0.41-5.66)) were most detected, followed by EBV (OR 1.37 (95%CI 0.33-4.88) and Parvovirus (OR 0.38 (95%CI 0,07-2.22). None of these viruses was more commonly detected in definite bacterial or definite viral cases. For the blood bacteria and fungi, none of the bacterial targets were more commonly detected in definite bacterial compared to definite viral. However, when comparing definite bacterial to all other phenotypes, 6 targets were more commonly detected in definite bacterial: pan bacterial (OR 6.52 (95%CI 2.24-18.99)), pan Staphylococcus (OR 7.00 (95%CI 1.69-29.08)), Enterobacter cloacae (OR 20.71 (95%CI 2.11-203.77)), Klebsiella pneumoniae (OR 13.49 (95%CI 1.20-151.95)), Enterobacteriales rpoB (OR 9.37 (95%CI 2.02-43.35)), and Enterobacteriales 16S (OR 14.10 (95%CI 2.50-79.41)).
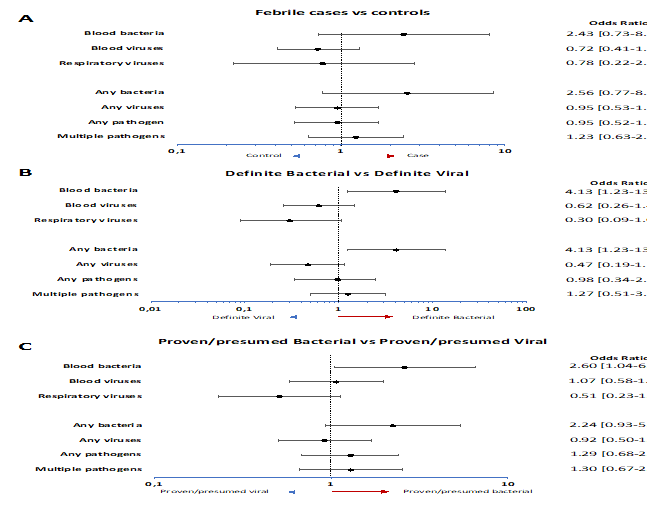
Figure 2: A) Odds ratios for molecular pathogen identification in groups, comparing febrile children and non-febrile controls. Dots show the mean and whiskers the 95% confidence intervals. Odds ratios >1 (right of dotted line; red arrow) indicate increased detection in cases. Odds ratios <1 (left of dotted line, blue arrow) indicate increased detection in controls.
B) Odds ratios for molecular pathogen identification in groups, comparing definite bacterial infection and definite viral infection. Dots show the mean and whiskers the 95% confidence intervals. Odds ratios >1 (right of dotted line; red arrow) indicate increased detection in definite bacterial cases. Odds ratios <1 (left of dotted line, blue arrow) indicate increased detection in definite viral cases.
C) Odds ratios for molecular pathogen identification in groups, comparing proven/presumed bacterial infection and proven/presumed viral infection. Dots show the mean and whiskers the 95% confidence intervals. Odds ratios >1 (right of dotted line; red arrow) indicate increased detection in proven/presumed bacterial cases. Odds ratios <1 (left of dotted line, blue arrow) indicate increased detection in proven/presumed viral cases.
CMT detection in febrile episodes of unknown etiology
In order to examine the potential utility of CMT results further, we assessed the detection rates of febrile episodes classified as unknown bacterial or viral infection. In 68 episodes CMT detected any pathogen (57.6%), of which 29 episodes with multiple pathogens detected (24.6%). Nine episodes had both bacterial and viral pathogens detected. In three episodes only bacterial pathogens were detected: Enterobacteriales, Mycoplasma pneumoniae, and pan Staphylococcus in one episode each. These were not detected on local microbiological investigations. In 57 episodes only viruses were detected, of which twelve episodes had multiple positive targets. In line with detection rates across different phenotypes, HHV7 (N=37), HHV6b (N=13) and EBV (N=8) were most commonly detected among episodes with only viral pathogens detected by CMT. Seven had parvovirus detected, five common cold viruses (three coronaviruses, one metapneumovirus and one parainfluenza type 2), and one each parechovirus and respiratory syncytial virus A.
Combining CMT and local diagnostics
When we combined local microbiological results and CMT, respiratory viruses were detected in 13.4% (N=6) and blood viruses in 44.5% (N=20) of definite bacterial febrile episodes. In a similar manner, in 16.2% of definite viral cases, evidence of bacteria or fungi was detected (N=8). Across all phenotypes evidence of viral pathogens was detected (Supplementary Figure 2). Across the cohort in 62 febrile episodes (18.4%) viruses and bacteria detected by local investigations also had a virus or bacteria detected by CMT.
When looking at matches in categories, i.e. gram-positive or gram-negative, all 17 bacterial matches were correct. In ten gram-negative cases, CMT matched a gram-negative pathogen, and likewise for the seven remaining gram-positive cases. However there were only three completely matched gram-negative cases (i.e. the exact same pathogen): one for Escherichia coli, one for Enterobacter cloacae, and one for Escherchia coli and Serratia marcescens in the same patient. Similarly there was one completely matched Staphylococcus aureus in the seven gram-positive matches. There were also three matches between coagulase negative Staphylococcus and the Pan Staphylococcal target. For viruses, there were 21 fully matched viral cases, and 24 viral mismatches. Of the viral mismatches 16 were HHV6 or HHV7, which are not always routinely screened for in local investigations.
As none of the CMT results clearly distinguished between definite bacterial and definite viral infection, these results could not be used to reclassify assigned phenotypes of the less certain phenotypes to definite bacterial or definite viral phenotypes.
Discussion
The international observational BIVA-HR study within the PERFORM study aimed to assess the etiology of febrile illness in immunocompromised children across Europe. This was performed by assessing current best practice of conventional diagnostics approaches and supplementary molecular diagnostic methods to detect bacteria and viruses in blood and respiratory samples. Despite thorough utilization of the methodologies, the vast majority of patients could not be assigned a bacterial or viral etiology. Based on current local best practice, only 13.4% (N=45) had definite microbiologically confirmed bacterial infection, and 11.0% (N=37) a viral infection without evidence of bacterial infection. The other 254 patients (75.6%) had varying degrees of certainty whether bacteria or viruses were the cause of febrile illness. There is a need for improved diagnostics as the burden of febrile illness is high and both bacterial and viral infection can cause significant morbidity and mortality, while it is also speculated that the majority of febrile illness in this group is self-limiting or due to non-infectious causes [5,17,18]. Applying CMT to confidently increase the proportion of febrile illness caused by definite bacterial or definite viral etiology was not feasible in this study, due to low detection rates of individual targets and subsequent capability to distinguish between definite bacterial and definite viral episodes. Nevertheless, when assessing the viral and bacterial targets as a whole, the detection of a bacterial target on CMT did seem to distinguish between bacterial and viral infection in proven cases. Higher detection of Enterobacter spp. targets when comparing definite bacterial cases to the other febrile cases, indicates that there might be a beneficial role for molecular testing for gram-negative pathogens. CMT increased the number of patients with detected viruses, although this was observed in all phenotypical categories, including the proven/presumed bacterial infections. There may be some overlap in the host response in bacterial and viral disease, and the detection of viruses in the blood does not fully exclude bacterial disease. This may imply that a significant proportion of immunocompromised children with febrile illness is co-infected with viruses. In this population, this further complicates accurate diagnosis, as viral presence may represent viral reactivation secondary to the underlying condition,not always causing fever and detectable for prolonged periods of time, thus not necessarily related to the febrile illness itself. Some of our findings will reflect the use of a variety of immunosuppressive or -modulating medication in our investigated patients, and therefore represent viral recrudescence, due to immune paresis by these medications. Thus, it might indicate both are involved in the patient’s illness, but biological significance of viral detection, mainly of herpes viruses, in relation to potential subsequent bacterial infection risk, remains unknown. Our inability to clearly differentiate patients between bacterial and viral infection on admission, is also reflected in clinical management. Most patients were prescribed antibiotics (89.3%), including those with a proven/presumed viral infection (75.3%).
Interestingly, across the cohort CMT detection rates of HHV7 (33.6% of episodes), HHV6b (12.5% of episodes) EBV (11.0% of episodes) was high, and these viruses combined made up 83.1% of all detected blood viruses (192/231). Similar proportions were observed in matched afebrile controls, with HHV6b even more likely to be detected in controls than in febrile episodes. This was intriguing, as these control patients did not show signs and symptoms of infection. Proportions of HHV7 and HHV6b were comparable in definite bacterial (20.5% and 15.9%) and definite viral (34.2% and 10.5%) episodes. It is known that herpes viruses, are latent in healthy individuals [39,40], hence, detection could represent bystander reactivation rather than being causative in the current febrile illness. Additionally, in approximately 1% of the general population, HHV-6 integration is inherited; this is known as inherited chromosomally integrated HHV-6 [41].In immunocompetent children, the commonality of HHV6b and HHV7 across the spectrum of febrile illness and healthy peers has been observed [42]. Further complicating interpretation, EBV, HHV, but also CMV and adenovirus are known to cause significant mortality and morbidity in certain immunocompromised patients, particularly in patients with T cell deficiencies and those post-hematopoietic stem cell transplantation [43-45]. In these patients, primary infection or viral reactivation is known to cause illness and they are PCR monitored throughout the course of transplant [46,47]. Yet, as these viruses are omnipresent in the studied population, we argue that the utility of testing for HHV6 and HHV7 on admission is of limited value. There are several potential explanations as to why we were unable to effectively distinguish bacterial and viral infections, even with the addition of a broad selection of pathogen targets with CMT. The febrile immunocompromised child represents a heterogenous group with a variety of underlying conditions and subsequent susceptibility to bacterial and viral pathogens, complicating uniform analysis. Yet this study does reflect the clinical presentation, as encountered by pediatricians across Europe working in emergency departments, more truthfully. Moreover, several common bacterial infections are localized, and are associated with bacteriaemia in more severe cases only. In this group, CMT in blood and conventional blood cultures might be equally sensitive, given the focus of infection is not the blood stream. Lastly, the frequency of detection of individual targets across the cohort was mostly low, complicating effective comparison, and those detected identified across the different phenotypical categories, and thus their importance is unclear. Our study indicates that the use of CMT might increase pathogen detection, but our findings do not support the clinical utility of CMT in clinical decision making about which febrile immunocompromised children definitely require antibiotics for potentially life-threatening bacterial infection or in whom they can safely be withheld. Detection of bacterial targets by molecular testing could be explored in further research as we observed that pan bacterial targets and a few gram-negative bacterial targets were more commonly detected in definite bacterial infection compared to the other patients in this cohort. This could potentially form a basis for risk stratification and treatment decisions as to whether or not to initiate antibiotics.
Strengths and limitations
Our study allowed us to analyse immunocompromised children across the European continent as encountered by clinicians in the emergency department. We used a specific set of viral, bacterial and fungal targets tailored for the immunocompromised child. We demonstrated the potential presence of viral and bacterial pathogens across the spectrum of febrile disease in this vulnerable population, as well as in matched controls. There are also several limitations. The heterogeneity of the cohort would facilitate generalizability of our results, although at present, we could not effectively substantiate the utility of CMT across the high-risk spectrum. We chose to look at this vulnerable population in the broadest sense, as there is a myriad of literature available to improve diagnostics for specifc subgroups of immunocompromised patients, especially oncological patients. Given that management pathways for these high-risk patients on presentation are very similar, use of CMT would be easiest to implement for this population as whole. Respiratory samples were not available for all participants, potentially reducing the detection of respiratory viruses, and subsequently underestimate the presence of respiratory pathogens in this cohort. Furthermore, low individual detection rates complicated effective comparison, and did not allow us to effectively analyze any quantitative CMT data, due to insufficient power. Therefore, we had to use qualitative PCR data only. This further complicated the interpretation of viral dection, as viral loads are used to aid the distinction between active infection and colonization, especially for latent viruses such as Herpesviridae. Due to the coronavirus disease 2019 pandemic, we had to limit the number of patients eligible for this study by limiting inclusion to the end of December 2019, to avoid pandemic-induced skewing of data. Interpretation is further complicated as there is no definite consensus on true positive bacterial infection, especially when bacterial cultures are negative, but clinically the suspicion of bacterial infection is high. Lastly, some patients received antibiotics in the community or prior to acquisition of the research samples, potentially leading to decreased number of confirmed bacterial cases in local and CMT tests.
Conclusion
This exploratory study shows that pathogen detection by molecular testing at present appears not be useful in differentiating bacterial and/or viral febrile illness from other causes of fever in high-risk patients at presentation to hospital. Consequently, it does not seem to further aid identifying the subset of immunocompromised patients in whom antibiotic treatment can be safely withheld. Therefore, we currently would not propose its use in the management of febrile illness at presentation, unless otherwise indicated. There appears to be no role for routine testing of HHV6b or HHV7 as these are present in similar proportion across the spectrum of febrile illness, and in afebrile children. Further studies are needed to identify if there is a role to test for (gram-negative) bacteria using CMT. Other, novel, approaches are needed to identify the population of immunocompromised febrile children in whom antibiotic treatment can be safely shortened or withheld.
Conflict of Interest
Authors Colin Fink and Marie Voice were employed by Micropathology Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Author Contributions
FvdV performed the statistical analyses and was the main author of this manuscript. PS and MV were responsible for selecting patients and conduct of the multiplex assays. ME conceptualized this project and supervised FvdV. EL, JH, EC, AK, AP, SP, UvB, LK, CV, NH, FMT, IRC, PA, LS, MT, IE, MP, MK, TK, DZ, AR, SY, MvdF, RdG, NS, WZ, ML, CF and ME were responsible for the conduct and sample collection for the PERFORM study at their respective sites. VW was responsible for sample management in the PERFORM study. TD maintained the clinical online database for the PERFORM study. All authors have read and provided valuable input during the writing of this manuscript. All authors agree with the submission of the final manuscript.
Funding
This project received funding under the European Union’s Horizon 2020 Research and Innovation programme under grant agreement numbers 668303 and 848196.
References
- Sands R, Shanmugavadivel D, Stephenson T, et al. Medical problems presenting to paediatric emergency departments: 10 years on. Emerg Med J 29 (2012): 379-382.
- Viscoli C, Varnier O, Machetti M. Infections in patients with febrile neutropenia: epidemiology, microbiology, and risk stratification. Clin Infect Dis 4 (2005): S240-245.
- Burcham MD, Cochrane AR, Jacob SA, et al. Emergency Department Chief Complaints Among Children With Cancer. J Pediatr Hematol Oncol 40 (2018): 445-449.
- Borensztajn DM, Hagedoorn NN, Carrol ED, et al. Febrile children with comorbidities at the emergency department - a multicentre observational study. Eur J Pediatr 181 (2022): 3491-500.
- Hakim H, Flynn PM, Knapp KM, et al. Etiology and clinical course of febrile neutropenia in children with cancer. J Pediatr Hematol Oncol 31 (2009): 623-629.
- Martinon-Torres F, Salas A, Rivero-Calle I, et al. Life-threatening infections in children in Europe (the EUCLIDS Project): a prospective cohort study. Lancet Child Adolesc Health 2 (2018): 404-414.
- Engert A, Balduini C, Brand A, et al. The European Hematology Association Roadmap for European Hematology Research: a consensus document. Haematologica 101 (2016): 115-208.
- Stryjewski GR, Nylen ES, Bell MJ, et al. Interleukin-6, interleukin-8, and a rapid and sensitive assay for calcitonin precursors for the determination of bacterial sepsis in febrile neutropenic children. Pediatr Crit Care Med 6 (2005): 129-135.
- van der Galiën HT, Loeffen EAH, Miedema KGE, et al. Predictive value of PCT and IL-6 for bacterial infection in children with cancer and febrile neutropenia. Support Care Cancer 26 (2018): 3819-3826.
- Xia T, Xu X, Zhao N, et al. Comparison of the diagnostic power of cytokine patterns and procalcitonin for predicting infection among paediatric haematology/oncology patients. Clin Microbiol Infect 22 (2016): 996-1001.
- Kitanovski L, Jazbec J, Hojker S, et al. Diagnostic accuracy of lipopolysaccharide-binding protein for predicting bacteremia/clinical sepsis in children with febrile neutropenia: comparison with interleukin-6, procalcitonin, and C-reactive protein. Support Care Cancer 22 (2014): 269-277.
- ?ahbudak Bal Z, Karada? Özdemir N, ?en S, et al. Diagnostic Accuracy of Interleukin-6, Interleukin-8, and Interleukin-10 for Predicting Bacteremia in Children with Febrile Neutropenia. Turk J Haematol 34 (2017): 254-257.
- van der Velden FJS, de Vries G, Martin A, et al. Febrile illness in high-risk children: a prospective, international observational study. Eur J Pediatr (2022).
- Aimoto M, Koh H, Katayama T, et al. Diagnostic performance of serum high-sensitivity procalcitonin and serum C-reactive protein tests for detecting bacterial infection in febrile neutropenia. Infection 42 (2014): 971-979.
- Baraka A, Zakaria M. Presepsin as a diagnostic marker of bacterial infections in febrile neutropenic pediatric patients with hematological malignancies. Int J Hematol 108 (2018): 184-191.
- Urbonas V, Eidukait? A, Tamulien? I. The diagnostic value of interleukin-6 and interleukin-8 for early prediction of bacteremia and sepsis in children with febrile neutropenia and cancer. J Pediatr Hematol Oncol 34 (2012): 122-127.
- Licciardello M, Pegoraro A, Cesaro S. Prophylaxis and therapy of viral infections in pediatric patients treated for malignancy. Pediatr Rep 3 (2011): e5.
- Lindblom A, Bhadri V, Soderhall S, et al. Respiratory viruses, a common microbiological finding in neutropenic children with fever. J Clin Virol 47 (2010): 234-237.
- Leigh S, Grant A, Murray N, et al. The cost of diagnostic uncertainty: a prospective economic analysis of febrile children attending an NHS emergency department. BMC Med 17 (2019): 48.
- Craig JC, Williams GJ, Jones M, et al. The accuracy of clinical symptoms and signs for the diagnosis of serious bacterial infection in young febrile children: prospective cohort study of 15 781 febrile illnesses 340 (2010): c1594.
- Randolph AG, McCulloh RJ. Pediatric sepsis: important considerations for diagnosing and managing severe infections in infants, children, and adolescents. Virulence 5 (2014): 179-189.
- Herberg JA, Kaforou M, Wright VJ, et al. Diagnostic Test Accuracy of a 2-Transcript Host RNA Signature for Discriminating Bacterial vs Viral Infection in Febrile Children. JAMA 316 (2016): 835-845.
- Srugo I, Klein A, Stein M, et al. Validation of a Novel Assay to Distinguish Bacterial and Viral Infections. Pediatrics 140 (2017).
- van der Velden FJS, Gennery AR, Emonts M. Biomarkers for Diagnosing Febrile Illness in Immunocompromised Children: A Systematic Review of the Literature. Front Pediatr 10 (2022): 828569.
- Arif T, Phillips RS. Updated systematic review and meta-analysis of the predictive value of serum biomarkers in the assessment and management of fever during neutropenia in children with cancer. Pediatr Blood Cancer 66 (2019): e27887.
- Mishra D, Satpathy G, Wig N, et al. Evaluation of 16S rRNA broad range PCR assay for microbial detection in serum specimens in sepsis patients. J Infect Public Health 13 (2020): 998-1002.
- Oberhettinger P, Zieger J, Autenrieth I, et al. Evaluation of two rapid molecular test systems to establish an algorithm for fast identification of bacterial pathogens from positive blood cultures. Eur J Clin Microbiol Infect Dis 39 (2020): 1147-1157.
- Poole S, Clark TW. Rapid syndromic molecular testing in pneumonia: The current landscape and future potential. J Infect 80 (2020): 1-7.
- Payne M, Champagne S, Lowe C, et al. Evaluation of the FilmArray Blood Culture Identification Panel compared to direct MALDI-TOF MS identification for rapid identification of pathogens. J Med Microbiol 67 (2018): 1253-1256.
- Shachor-Meyouhas Y, Sprecher H, Moscoviz D, et al. Molecular-based diagnosis of bacteremia in the setting of fever with or without neutropenia in pediatric hematology-oncology patients. J Pediatr Hematol Oncol 35 (2013): 500-503.
- Santolaya ME, Farfan MJ, De La Maza V, et al. Diagnosis of bacteremia in febrile neutropenic episodes in children with cancer: microbiologic and molecular approach. Pediatr Infect Dis J 30 (2011): 957-961.
- Idelevich EA, Silling G, Niederbracht Y, et al. Impact of multiplex PCR on antimicrobial treatment in febrile neutropenia: a randomized controlled study. Med Microbiol Immunol 204 (2015): 585-592.
- Lamoth F, Jaton K, Prod'hom G, et al. Multiplex blood PCR in combination with blood cultures for improvement of microbiological documentation of infection in febrile neutropenia. J Clin Microbiol 48 (2010): 3510-3516.
- Ray ST, Drew RJ, Hardiman F, et al. Rapid Identification of Microorganisms by FilmArray Blood Culture Identification Panel Improves Clinical Management in Children. Pediatr Infect Dis J 35 (2016): e134-138.
- Lucignano B, Ranno S, Liesenfeld O, et al. Multiplex PCR allows rapid and accurate diagnosis of bloodstream infections in newborns and children with suspected sepsis. J Clin Microbiol 49 (2011): 2252-2258.
- Dark P, Blackwood B, Gates S, et al. Accuracy of LightCycler((R)) SeptiFast for the detection and identification of pathogens in the blood of patients with suspected sepsis: a systematic review and meta-analysis. Intensive Care Med 41 (2015): 21-33.
- Consortium P. Personalised Risk assessment in Febrile illness to Optimse Real-life Management across the European Union - Clinical Protocol (2016).
- Nijman RG, Oostenbrink R, Moll HA, et al. A Novel Framework for Phenotyping Children With Suspected or Confirmed Infection for Future Biomarker Studies. Front Pediatr 9 (2021): 688272.
- Pass RF. HHV6 and HHV7: persistence and vertical transmission. J Pediatr 145 (2004): 432-435.
- Thorley-Lawson DA, Hawkins JB, Tracy SI, et al. The pathogenesis of Epstein-Barr virus persistent infection. Curr Opin Virol 3 (2013): 227-232.
- Pantry SN, Medveczky PG. Latency, Integration, and Reactivation of Human Herpesvirus-6. Viruses 9 (2017).
- Shah P, Voice M, Calvo-Bado L, et al. Relationship Between Molecular Pathogen Detection and Clinical Disease in Febrile Children Across Europe. Preprints with The Lancet (2022).
- Annaloro C, Serpenti F, Saporiti G, et al. Viral Infections in HSCT: Detection, Monitoring, Clinical Management, and Immunologic Implications. Front Immunol 11 (2020): 569381.
- Ljungman P. Molecular monitoring of viral infections after hematopoietic stem cell transplantation. Int J Hematol 91 (2010): 596-601.
- Styczynski J, van der Velden W, Fox CP, et al. Management of Epstein-Barr Virus infections and post-transplant lymphoproliferative disorders in patients after allogeneic hematopoietic stem cell transplantation: Sixth European Conference on Infections in Leukemia (ECIL-6) guidelines. Haematologica 101 (2016): 803-811.
- Hiwarkar P, Gaspar HB, Gilmour K, Jagani M, Chiesa R, Bennett-Rees N, et al. Impact of viral reactivations in the era of pre-emptive antiviral drug therapy following allogeneic haematopoietic SCT in paediatric recipients. Bone Marrow Transplant 48 (2013): 803-808.
- Laberko A, Gennery AR. Clinical considerations in the hematopoietic stem cell transplant management of primary immunodeficiencies. Expert Rev Clin Immunol 14 (2018): 297-306.
ORCID IDs
Fabian JS van der Velden 0000-0001-7864-4074
Priyen Shah 0000-0001-9164-8862
Emma Lim 0000-0002-5403-3065
Jethro Herberg 0000-0001-6941-6491
Victoria Wright 0000-0001-7826-1516
Enitan D. Carrol 0000-0001-8357-7726
Stéphane Paulus 0000-0002-0703-9114
Ulrich von Both 0000-0001-8411-1071
Laura Kolberg 0000-0002-3311-8254
Clementien L Vermont 0000-0002-9196-6710
Nienke N Hagedoorn 0000-0001-9237-4904
Federico Martinón-Torres 0000-0002-9023-581X
Irene Rivero Calle 0000-0002-3678-9264
Philipp KA Agyeman 0000-0002-8339-5444
Luregn J Schlapbach 0000-0003-2281-2598
Maria Tsolia 0000-0003-2485-4409
Irini Eleftheriou 0000-0002-5093-8502
Marko Pokorn 0000-0002-2341-2791
Mojca Kolnik 0000-0003-4194-7295
Taco W Kuijpers 0000-0002-7421-3370
Dace Zavadska 0000-0003-4892-3763
Aleksandra Rudzate 0000-0003-4885-0031
Werner Zenz 0000-0002-6218-3143
Shunmay Yeung 0000-0002-0997-0850
Michiel van der Flier 0000-0003-1296-0159
Michael Levin 0000-0003-2767-6919
Colin Fink 0000-0002-8008-5870
Marieke Emonts 0000-0002-2822-3527


 Impact Factor: * 5.3
Impact Factor: * 5.3 Acceptance Rate: 75.63%
Acceptance Rate: 75.63%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 