Prevalence of Respiratory Pathogens in COVID Patients
Johnny Michel1,2, Maria-Alexandra Stoica3, Myriam Aouiti-Trabelsi2, Fabienne DE Oliveira3, Eva Hong2, Luc-Marie Joly1, Ala-Eddine Deghmane2, Jean-Christophe Plantier3*, Muhamed-Kheir Taha2*
1Emergency Department, Rouen University Hospital, F 76000, Rouen, France
2Institute Pasteur, Université Paris Cité, Invasive Bacterial Infections Unit, F 75115, Paris, France
3Microbiology Department, Rouen University Hospital, F 76000, Rouen, France
*Corresponding author: Jean-Christophe Plantier, Microbiology Department, Rouen University Hospital, F 76000, Rouen, France.
Muhamed-Kheir TAHA, Institute Pasteur, Invasive Bacterial Infections Unit, F 75115, Paris, France.
Received: 26 July 2023; Accepted: 01 August 2023; Published: 06 October 2023
Article Information
Citation: Johnny Michel, Maria-Alexandra Stoica, Myriam Aouiti-Trabelsi, Fabienne DE Oliveira, Eva Hong, Luc-Marie Joly, Ala-Eddine Deghmane, Jean-Christophe Plantier, Muhamed-Kheir Taha. Prevalence of Respiratory Pathogens in COVID Patients. Journal of Biotechnology and Biomedicine. 6 (2023): 450-459.
DOI: 10.26502/jbb.2642-91280107
View / Download Pdf Share at FacebookAbstract
Background: Management of a novel respiratory virus causing severe pneumonitis included the use of antibiotics to prevent bacterial co-infections and secondary infections. However, the impact of this antibiotic use on the selection of resistant bacterial isolates needs to be evaluated.
Methods: We conducted a single-center retrospective study from November 14, 2020 to December 31, 2021 to assess the prevalence of several members of the nasopharyngeal microbiota from PCR-positive SARS-CoV-2 subjects. The study population corresponded to 1030 nasopharyngeal swabs positive for SARS-CoV-2 at the university hospital of Rouen site in symptomatic patients aged 16 years and older. Real-time PCR was used to detect the presence of Haemophilus influenzae, Streptococcus pneumonia, Neisseria meningitidis and influenza A virus. An analysis of the ftsI gene was further used to analyze beta-lactam resistance in H. influenzae.
Results: The results reveled less than expected carriage rate with 5% for H. influenzae, 1.2% for N. meningitidis and 3.7% for S. pneumoniae and an absence of influenza A. On the other hand, there was a significant difference (p<0.01) between the "carriage" and "no carriage" groups on age, sex, oxygen therapy and orotracheal intubation, implying a more severe evolution of the COVID-19 in carriers. Analysis of the ftsI gene reveals 26% of predicted resistance to amoxicillin without resistance to third generation cephalosporins.
Conclusions: COVID-19 pandemic has disrupted bacterial and viral epidemiology, leading to lower circulation of several respiratory pathogens.
Keywords
<p>COVID; Viral epidemiology; Bacterial infection; Pneumoniae; Virus</p>
Article Details
Introduction
The new coronavirus-2019 (COVID-19) infectious disease, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has traumatized the world with a pandemic that is still ongoing with a seventh wave for the summer of 2022 in France. In addition to non-pharmaceutical control measures, the arrival of a new respiratory agent can impact the nasopharyngeal microbiota. The major nasopharyngeal agents identified with SARS-CoV-2 include bacteria such as Streptococcus pneumoniae, Staphylococcus aureus, Klebsiella pneumoniae, and Haemophilus influenzae [1–3] as well as viral agents. Some of these agents can be the cause of invasive bacterial infections. Changes induced by SARS-CoV-2 may also synergize the pathogenesis of these agents. In addition, co-infecting microbiota may use novel strategies to evade host defense mechanisms by altering innate and adaptive immune responses, which may further exacerbate the pathogenesis of SARS-CoV-2 [3]. A better understanding of co-infections in COVID-19 is essential for effective patient management, treatment and containment of SARS-CoV-2. Thus, identification of possible SARS-CoV-2 co-pathogens may help providing better management of these cases of mixed infections [4]. Smith et al [5] reported in March 2022 that SARS-CoV-2 infection increases susceptibility to secondary bacterial infection. These co-infections remain uncommon, but the incidence of ICU-acquired infections is very high with a poorer prognosis [6,7]. An assessment and a better understanding of respiratory pathogens that are co-infecting or superinfecting with SARS-CoV-2 is necessary to optimize the management of COVID-19 patients and to improve antibiotic management. Humans can be asymptomatic carriers of several potentially invasive bacterial species. These include S. pneumoniae (Sp), Neisseria meningitidis (Nm) and H. influenzae (Hi). These bacteria can be the cause of secondary invasive infections following a viral respiratory disease, such influenza A virus infections [8]. Several studies suggest a decline in invasive bacterial infections due to N. meningitidis, H. influenzae and S. pneumoniae during the COVID-19 pandemic [9–11]. This decrease can be explained on the one hand by the implementation of barrier non-pharmaceutical measures with social distancing and the use of masks but also by the decrease of viral agents, such as influenza, and thus a decrease of secondary bacterial infections [12].
The prevalence of carriage of these bacteria varies with age, with frequent carriage of H. influenzae and S. pneumoniae in children up to 2 years of age (approximately 43% and 32% respectively) [13]. Carriage of N. meningitidis is rare at this age but frequent between 16 and 24 years of age (about 25%) [14]. In addition, increasing proportions of H. influenzae isolates (mostly from respiratory sites) are resistant to ampicillin and to amoxicillin that is mediated by the acquisition of a beta-lactamase and/or modification of Penicillin-Binding Protein 3 (PBP3) by mutation of the ftsI gene encoding the PBP3 involved in the beta lactamase-independent resistance to beta-lactams [15,16]. In addition, isolates resistant to third generation cephalosporins are also emerging through mutations in the ftsI gene [15]. The objective of this work is to evaluate the prevalence of three potentially invasive bacterias and influenza A virus in a COVID-19 positive population, as well as to analyze the ftsI-mediated resistance of H. influenzae to beta-lactams.
Patients, Materiel and Methods
Patients and samples
This is a single-center retrospective descriptive study at the University Hospital Center (C.H.U) of Rouen to evaluate the prevalence of bacterial and viral carriage in symptomatic patients of COVID-19, over the period from November 14, 2020 to December 31, 2021, corresponding to the period after first and second waves in France. The study population corresponded to PCR-positive nasopharyngeal swabs taken for the detection of SARS-CoV-2 at the C.H.U of Rouen in symptomatic patients aged 16 years and older. The result was positive for the detection of SARS-CoV-2 by molecular methods performed at the Rouen University Hospital. For the homogeneity of the analysis, we selected only samples from the period 14/11/2020 to 31/12/2021 from patients who attended the adult emergency department, from the different intensive care units deployed and from the main COVID units of the hospital. The patients could be divided into 3 groups of orientation management: "Ambulatory", "Convention Care Unit (CCU)" and "Intensive Care Unit (ICU)".
In accordance with the law regarding the General Data Protection Regulation and applicable regulations, the retrieved data were anonymized, and no step in this retrospective study infringed on patient anonymity, respect, or integrity. Data were obtained from the patients' electronic medical records in the hospital database. Moreover, this study was approved by the Ethics Committee for Research on Existing Data of the C.H.U of Rouen. The need for informed consent was waived by the ethics committee/Institutional Review Board of [the Comité d'Ethique pour la Recherche sur Données Existantes et/ou hors loi Jardé du Centre Hospitalier Universitaire de Rouen (France), because of the retrospective nature of the study. The study was performed in compliance with the latest version of the Declaration of Helsinki dated October 2013. On the period from November 14, 2020 to December 31, 2021, we obtained 1030 primary samples, that were stored at -80°C. The samples corresponded to the all-available samples that were extracted at the laboratory of the C.H.U of Rouen using MGIEasy Magnetic Beads Virus DNA/RNA Extraction Kit on MGI SP-960 instruments (MGI Tech Co. Shenzhen, China) according to the manufacturer’s recommendations and then sent to the Pasteur Institute in Paris for analysis. Clinical data were obtained from the electronic medical records of the patients in the database of the C.H.U of Rouen. Data collection included age, sex, date of collection, care unit of collection, ambulatory, conventional care unit or intensive care unit orientation, presence of oxygen therapy and its degree (simple oxygen therapy, high flow oxygen therapy (HFO) Or Orotracheal Intubation (OTI)), survival to stay for COVID-19 infection.
Molecular analysis
The molecular detection was performed on the basis of PCR amplification of specific genes for the three bacterial species (Nm, Hi and Sp) and for influenzae A virus (IAV) [17,18]. This allows the identification of H. influenzae (hpd) and the determination of whether the strain is capsulated (bexD). The PCR Ct values of 35 and lower were considered positive and Ct values >40 were considered negative. For samples Ct >35 and <40, an additional PCR with the ompP2 gene (H. influenzae) was performed based on the ompP2 gene (Table 1). As for the screening for N. meningitidis and S. pneumoniae, identification was performed by PCR amplification of the sodC and lytA genes respectively, with identification of groupable meningococci with the ctrA gene. For the influenza A virus, we performed a generic RT-PCR of the M gene (PCR InfA) which codes for the M matrix protein. The primers used were summarized in Table 1. This first screening real-time PCRs for the detection of the targeted agents were performed on a StepOnePlus (Applied Biosystems™).
|
Pathogen |
Target |
Primers/Probe |
Nucleotide sequence (5’-3’) |
|
Hi |
hpd |
Forward |
-5’GGTTAAATATGCCGATGGTGTTG-3’ |
|
Reverse |
5-’Tgcatctttacgcacggtgta-3’ |
||
|
Probe |
FAM-5’-TTGTGTACACTCCGTTGGTAAAAGAACTTGCAC-3’ |
||
|
Hi |
bexD |
Forward |
5’-GCAGTGGTATTACGCTTGCTG-3’ |
|
Reverse |
5’-ATACCCCTTAGCCGCCCA-3’ |
||
|
Probe |
VIC-5’-ACGCGTTCTGACCCACGAGGTGT-3’ |
||
|
Hi |
ompP2 |
Forward |
5’-GGTGCATTCGCAGCTTCAG-3’ |
|
Reverse |
5’-GATTGCGTAATGCACCGTGTT-3’ |
||
|
Probe |
FAM-5’-TTGTTTATAACAACGAAGGGACTAACGT-3’ |
||
|
Nm |
sodC |
Forward |
5’-GCACACTTAGGTGATTTACCTGCAT-3’ |
|
Reverse |
5’-CCACCCGTGTGGATCATAATAGA-3’ |
||
|
Probe |
FAM-5’-CATGATGGCACAGCAACAAATCCTGTTT-3’ |
||
|
Nm |
ctrA |
Forward |
5’-GCTGCGGTAGGTGGTTCAA-3’ |
|
Reverse |
5’-TTGTCGCGGATTTGCAACTA-3’ |
||
|
Probe |
FAM-5’-GTGCAGGATACGAATGTGCAGCTGAC-3’ |
||
|
Sp |
lytA |
Forward |
5’-ACGCAATCTAGCAGATGAAGCA-3’ |
|
Reverse |
5’-TCGTGCGTTTTAATTCCAGCT-3’ |
||
|
Probe |
VIC-5’-TGCCGAAAACGCTTGATACAGGGAG-3’ |
||
|
IAV |
infA |
Forward |
5’-GACCRATCCTGTCACCTCTGAC-3’ |
|
Reverse |
5’-AGGGCATTYTGGACAAAKCGTCTA-3’ |
||
|
Probe |
FAM-5’-TGCAGTCCTCGCTCACTGGGCACG-3’ |
||
|
Hi |
ftsI external |
Forward |
5’-GGAAGTGTTAGCTATGGCGAC-3’ |
|
Reverse |
5’-GGGCAGAAACCGCACCACCAT-3’ |
||
|
Forward |
5’-GTTTTCCCAGTCACGACGTTGTAG |
||
|
Hi |
ftsI internal |
TTAATGCGTAACCGTGCAATTAC-3’ |
|
|
Reverse |
5’-TTGTGAGCGGATAACAATTTCA |
||
|
CCACTAATGCATAACGAGGATC-3’ |
|||
|
FAM fluorochrome: absorbance at 494nm and mission at 520nm |
|||
Table 1: Nucleotide sequences of PCR primers and probe for the different screening genes.
For samples that were positive for Hi, we next performed a second PCR on the ftsI gene using an end-point nested PCR (Table 1). The amplicons were then analyzed by 1% agarose gel electrophoresis. After purification by gel filtration on Sepharose CL-6B column (VMR). The obtained products were Sanger-sequenced and the sequences were aligned using multiple sequence alignment CLUSTALW and used to build up network and phylogram that were generated using Splitstree version 4.0 [19]. ftsI allele identification was performed using BIGSdb tools on the PUBMLST database https://pubmlst.org/organisms/haemophilus-influenzae [20].
Statistical analysis
GraphPad Prism Software 9.3.1 (San Diego, California USA, www.graphpad.com) was used to describe quantitative variables that were expressed as mean and standard deviation or median and interquartile range if their distribution did not follow a normal distribution. The qualitative variables were expressed as numbers and percentages. Patients were classified according to the variable "orientation" and compared for the qualitative variables using the chi-square test or Fisher's exact test when the conditions were not met. Comparison of quantitative variables was performed using the Kurskal-Walis test for a significant difference between groups and supplemented by a Bonferoni-adjusted Dunn test to identify different groups in case of a significant difference. Patient’s data were also compared according to the detection/non-detection of at least one of the three bacterial agents. For quantitative variables, we used a student’s t-test completed by a Wilcoxon/Mann-Whitney test in front of a non-normality of distribution verified by a Shapiro-Wilk test. For qualitative variables, we used a Fisher exact test. Results were expressed as odds ratios. Data analysis was performed using R® software (version 2022.02.2+485) at a consenting alpha risk of 5% (with Bonferoni correction for multiple comparisons).
Results
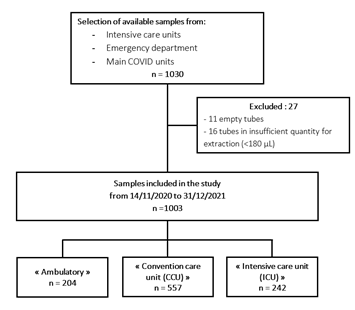
Figure 1: Flow chart N.
Patient’s characteristics
The study population included 1003 positive for SARS-COV-2 samples corresponding to 1003 patients (Figure 1). Patient characteristics were summarized in Table 2. Patients were classified according to increasing severity. The "ambulatory" group include symptomatic patients who were seen at the adult emergency department and discharged without hospitalization. The "conventional care unit" group include symptomatic patients requiring hospitalization without intensive care. The "intensive care unit" group included patients requiring intensive care, mainly for high flow oxygen therapy or intubation due to respiratory distress. The mean age of our population was 66 years with extremes of age from 16 to 104 years. The proportion of males and females in the total population yielded a sex ratio of 1.22 (Table 2).
We first analyzed the global distribution of different variables scored in this study among the three groups of patients as defined in the Method section. The details of the comparative statistics on the referral group are summarized in Table 2. There was a significant difference in the survival criterion between the three groups (p < 0.0001), more marked in the outpatient group than in the inpatient and intensive care groups, with a survival rate of 100% in the outpatient group (ambulatory group), 78.8% in the inpatient group (CCU group) and 76.5% in the intensive care group (ICU group) respectively. There was also a significant difference on the "sex" criterion for all three groups (p < 0.001), more pronounced with a multiple Fisher test between the CCU and ICU groups (p < 0.001), and between the ambulatory and ICU groups (<0.05), but no significant difference between ambulatory patients and those hospitalized in conventional care. The use of oxygen therapy technique was also significantly different between the three observed groups (p < 0.0001).
|
Patients |
Total |
Ambulatory |
CCU |
ICU |
p-value |
|
N = 1003 (%) |
204 (20.4) |
557 (55.5) |
242 (24.1) |
||
|
Median age [IIQ] |
68 [52-83] |
47 [30.75-66] |
79 [65-87] |
62 [51.25-70] |
< 0.0001 |
|
Maximum age |
104 |
104 |
100 |
85 |
- |
|
Minimum age |
16 |
16 |
17 |
21 |
- |
|
Survival |
828 (82.6) |
204 (100) |
439 (78.8) |
185 (76.5) |
< 0.0001 |
|
No oxygen therapy |
352 (35) |
203 (99.5) |
146 (26.2) |
3 (1.2) |
< 0.0001 |
|
Oxygen therapy |
426 (42.5) |
1 (0.5) |
411 (73.8) |
14 (5.8) |
|
|
High flow oxygen therapy |
96 (9.6) |
0 |
0 |
96 (39.7) |
|
|
Tracheal intubation |
129 (12.9) |
0 |
0 |
129 (53.3) |
|
|
Male |
552 (55) |
110 (53.9) |
284 (51) |
158 (65.3) |
< 0.001 |
|
Haemophilus influenzae |
7 (3.4) |
27 (4.8) |
16(6.6) |
0.3 |
|
|
Neisseria meningitidis |
0 (0) |
6 (1.1) |
6 (2.5) |
0.05 |
|
|
Streptococcus pneumoniae |
5 (2.5) |
21 (3.8) |
11 (4.5) |
0.5 |
|
|
Influenzae A |
0 |
0 |
0 |
- |
Table 2: Population characteristics and comparative statistics between orientation groups.
PCR-based detection of microbial agents
PCR-based detection of Hi using hpd and ompP2 genes allowed the detection of 50 Hi-positive samples with an average Ct value of 32 accounting for 5.0% of all the 1003 tested samples. All the 50 samples were negative for bexD gene suggesting that they corresponded to non-capsulated (non-typeable) isolates. PCR-based detection of Nm was performed on the basis of the detection of sodC gene that yielded 12 samples positive for the presence of N. meningitidis, with an average Ct value of 32 and corresponding to 1.2% of all the 1003 tested samples PCR analysis using ctrA gene identified 6 ctrA positive samples suggesting that they correspond to groupable meningococci but complete analysis could not be performed due to lack of sufficient extract. PCR-based detection of S. pneumoniae used with the lytA gene revealed 37 positive samples with an average Ct value of 33 and corresponding to 3.7% of all the 1003 tested samples. Finally, no sample was positive for the presence of influenza A virus. Double bacterial carriage was observed for 16 samples. No sample was positive for the presence of the 3 bacterial agents tested. A temporal distribution of bacterial carriage is shown in Figure 2. The prevalence curves as a function of time overlap for H. influenzae and S. pneumoniae carriage. They appear to follow the trend in the frequency of COVID-19 infection. A concomitant increase of COVID-19 cases as well as the detection of the three bacterial agents was observed since November 2021 after easing from the two-periods of lockdowns in France that were implemented during the study period. The first period, from October 30 to December 15, 2020, was partially included in our study, and the second period was from April 3 to May 3, 2021 (Figure 2). There was no significant difference between the referral groups on the carriage of H. influenzae, N. meningitidis or S. pneumoniae.
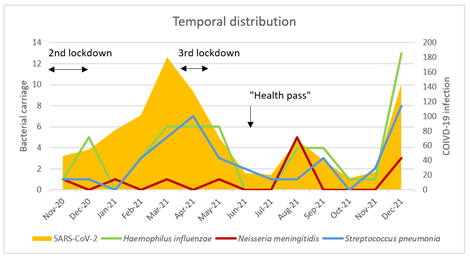
Figure 2: Temporal distribution of SARS-CoV-2 prevalence as well as carriage of different germs and influenza in the population of the study.
Association of the presence of the three bacterial agents with the groups of patients
We next compared the distribution of the variables scored in this study between one group called "carriage" group, which includes patients with samples positive for the presence of at least one of the three bacteria in nasopharyngeal carriage, and a "no carriage" group corresponding to patients with samples for whom none of the 3 bacteria tested was detected (Table 3). The "carriage" group is younger (p<0.01) than the “non-carriage” and includes more males (p<0.01).
|
Patients |
Bacterial carriage |
No carriage |
|
|
p-value |
||
|
N=1003 |
N=83 (8,3%) |
N=920 (91,7%) |
|
|
|||
|
|
Median |
[IIQ] |
Median |
[IIQ] |
|
|
|
|
Age |
64 |
[46-72] |
69 |
[53-83] |
< 0,01 |
||
|
|
Number |
(%) |
Number |
(%) |
OR |
[IC 95%] |
|
|
Sex (male) |
59 |
(71,1) |
493 |
(53,6) |
2.1 |
[1,3-3,6] |
< 0,01 |
|
Survival |
74 |
(89,2) |
754 |
-82 |
0,55 |
[0,2-1,1] |
0,13 |
|
No oxygen therapy |
20 |
(24,1) |
332 |
(36,1) |
0,56 |
[0,3-0,9] |
0,03 |
|
Oxygen therapy |
38 |
(45,8) |
388 |
(42,2) |
1,16 |
[0,7-1,9] |
0,56 |
|
HF oxygen therapy |
6 |
(7,2) |
90 |
(9,8) |
0,72 |
[0,2-1,7] |
0,56 |
|
Tracheal ntubation |
19 |
(22,9) |
110 |
-12 |
2,18 |
[1,2-3,8] |
< 0,01 |
|
All oxygen therapy cases |
63 |
-76 |
588 |
-64 |
1.8 |
[1,0-3,2] |
0.04 |
|
Amubulatory |
10 |
-12 |
194 |
(21,) |
0,51 |
[0,2-1,0] |
0,06 |
|
Hospitalization |
47 |
(56,6) |
510 |
(55,4) |
1,04 |
[0,6-1,7] |
0,90 |
|
Intensive care |
26 |
(31,3) |
216 |
(23,5) |
1,48 |
[0,9-2,5] |
0,11 |
|
All hospitalization cases |
73 |
-88 |
726 |
-76 |
1.9 |
[1,0-4,3] |
0.07 |
Table 3: Comparative statistics between patients with and without bacterial carriage.
There was a significant difference in the need for oxygen therapy, with the "non-carriage" group seeming to have less need for oxygen therapy (p<0.05). Patients requiring oxygen therapy were significantly more frequent among carriage (p = 0.04) and this was mainly observed among patient with intubation (p<0.01). High flow and simple oxygen therapy was not significantly different.
Concerning the orientation of the patient, there was no significant difference between the two groups.
Among the 19 patients intubated in the "carriage" group, 16 samples were positive for H. influenzae, 11 for S. pneumoniae and 6 for N. meningitidis. In this sub-population, 8 patients presented X-ray and bacteriological findings suggesting a pulmonary bacterial superinfection with H. influenzae, such as pneumopathy may have been acquired under mechanical ventilation, with a possible passage of the bacterial carriage during intubation. All these patients were treated with beta-lactams.
Prediction of susceptibility to beta-lactam for Hi-positive samples
The use of beta lactams to treat patients suffering from a suspected pulmonary bacterial superinfection mainly with H. influenzae led us to focus on genetic typing of beta lactam susceptibility of H. influenzae positive samples. We therefore analyzed the sequence of the ftsI gene to predict beta-lactamase independent resistance to amoxicillin and third generation cephalosporins (3GC). The sequences of fstI gene were successfully obtained from 621 bp ftsI PCR-amplified fragments from all the 50 Hi positive samples. These DNA sequences allowed the identification of 29 different ftsI alleles and were named ftsIn where n is the allele number. Alleles 312, 313, and 314 were new alleles identified in our study and were added to the PUBMLST database. The 29 alleles were grouped into three groups 1 to 3 according to the already described scheme based on to the presence/absence of mutations in ftsI that were suggested to be involved in beta-lactam resistance [15] (Table 4). The allele frequencies among samples varied from 1 to 6 samples with 7 alleles that were represented by more than one sample (ftsI4, ftsI8, ftsI10, ftsI27, ftsI50, ftsI193 and ftsI314). Concerning the frequency of the groups of beta-lactam resistance, we note a prevalence for alleles of group 1, that do not harbor any mutations in the critical region of PBP3. This group was represented by 10 alleles found in 25 samples (50% of all the 50 Hi-positive samples). Alleles of Group 2, that do not harbor mutations associated with resistance to beta lactams but present other polymorphic sites in the critical region of PBP3, were represented by 7 alleles that were found in 12 samples (24% of all the 50 Hi-positive samples). Alleles of Group 3, that harbor mutations associated with resistance to beta lactams in the critical region of PBP3, were represented by 12 alleles that were found in 13 samples (26% of all the 50 Hi-positive samples). The alleles are correlated to beta-lactamase negative ampicillin resistance [15] (Table 4). No allele belonging to group 4, correlated to the resistance to 3GC and harboring four key mutations (D350N, S357N, M377I and S385T) was detected [15]. The phylogenetic tree drawn by SplitsTree4 based on the CLUSTALW alignment of amino acid sequences deduced from the 29 ftsI alleles (Fig.3) clearly showed the clustering of the 29 deduced amino sequences into the 3 groups. One allele in group 2 (ftsI302) was distantly separated from the other group 2 alleles and seems to be also shared by Haemophilus parainfluenzae (on the data base pubmlst.org/organisms/haemophilus-influenzae access on the 08/11/2022) [15].
|
Group* |
Alleles (n° of isolates if >1) |
Mutation associate with amoxicillin resistance (N526K)* |
Mutations associate with third generation cephalosporine resistance (D350N, S357, M377I, S385T)* |
Other mutations |
|
1 |
4 (2), 8(5), 10(5), 15, 27(6), 37, 63, 70, 156, 193 (2) |
No |
No |
No |
|
2 |
39, 50(5), 55, 97, 116, 302 and 314 (2) |
No |
No |
Yes |
|
3 |
1, 5, 20, 21, 48(2), 122, 142, 153, 266, 272, 312, 313 |
Yes |
No |
Yes |
|
*The groups and key mutations for resistance were according to Deghmane et al (15). Alleles (55 and 39); (5, 122) (20, 142) and (153, 313) showed identical amino-acid sequences |
||||
Table 4: Mutations in the ftsI gene
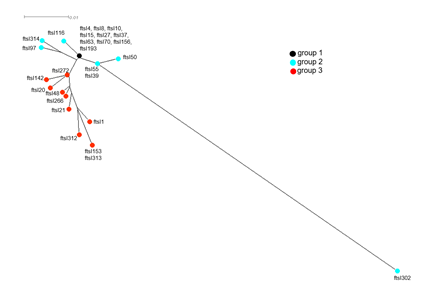
Figure 3: CLUSTALW-based ftsI phylogenetic tree multiple alignment of amino acid sequences deduced from the DNA sequences of all ftsI alleles defined among the invasive isolates in this study. The tree was visualized by SplitsTree4 as described in the Methods section.
Discussion
This is a single-center, retrospective study. The population was selected based on PCR-confirmed SARS-CoV-2 infection in nasopharyngeal swabs. It is worthy to note that we used PCR to detect the bacterial agents that is more sensitive than culture in detecting the presence of bacteria [21]. Our data did not reveal significant differences in term of carriage of Hi, Nm or Sp according to the clinical classification of COVID-19 cases. However, one element that stands out from our study is that bacterial carriage of at least one of these three agents seems to indicate a greater need for oxygen therapy in COVID-19 pathology and management as well as a greater risk of intubation in severe pathology (p<0.001) and an overall difference with oxygen therapy requirement among carriers. One important observation in our study is the low prevalence of the three bacterial agents, Hi, Nm and Sp (between 1 and 5%). Carriage rate varies with age and settings for the three agents with an expected carriage rate in adult population for the three agents around 10% [14,22,23]. Our study period concerns a population from November 2020 to December 2021, after the first and second waves of COVID-19, and therefore after the implementation of barrier and containment measures in France. However, further stratification of our data was not pertinent due to low number of isolates. Containment measures would impact on the transmission of the three respiratory bacterial agents and therefore carriage rates may be expected to decrease. Additionally, this redaction may be due to the presence of a new viral agent (SRAS-CoV-2) in the pharyngeal microbiota. Of interest, a decrease in invasive infections has already been described in several previous studies, notably for S. pneumoniae [24] but also for H. influenzae [25] and N. meningitidis [11]. This effect is most likely related to the introduction of containment policies during the COVID-19 pandemic, reducing transmission, mainly respiratory droplets with wearing of masks and social distancing [10]. However, the study by Danino et al in 2021 [12], suggests that the decrease of S. pneumoniae infections is less related to reduced bacterial carriage than to the decrease of viral infections (such as influenza viruses and respiratory syncytial viruses) , which usually promote secondary invasive bacterial infections. In agreement with this hypothesis, we did not detect the presence of influenza A virus in our samples of SARS-CoV-2 patients.
A perennial model of decline in pneumococcal infections [26] suggests a reduction in pneumococcal carriage and invasive infections for up to 5 years after the end of the containment period. This result suggests monitoring of carriage to confirm this impact after removal of these measures. However, several other factors may be involved such as the reduction of natural immunity in the population due to the diminution of the circulation of these bacterial species that can be synergized by reduced vaccine uptake during last two years [27,28]. Such reduction may be responsible for an ‘immunity-gap” that can promote a rebound in these invasive bacterial infections [29]. Indeed, a rapid rebound seems to occur for N. meningitidis in England [30] and France [31] since the end of 2021. This is also in agreement with our results of the rapidly increased circulation of all three bacterial agents since November 2021 (Figure 2). ftsI phylogenic tree highlights the heterogeneity of the alleles in our H. influenzae positive samples. Group 3 alleles (n=12; 13 isolates 26%) correlated with amoxicillin and penicillin resistance. This result is similar to the percentage of 24% that was previously reported in France for beta-lactamase negative ampicillin resistant Hi isolates [15]. However, we did not perform PCR-based detection of genes encoding beta-lactamases as this search will not allow to attribute the detected beta-lactamase genes to a bacterial species. It is interesting that we did not detect group 4 ftsI (associated to 3GC resistance). However, we defined new fstI alleles for which no phenotypic data are yet available. Alternatively, new mutations may be also involved in resistance to 3GC and remained to be identified. One limitation of our study is the lack of a "SARS-CoV-2 negative" control group and thus preventing to evaluate the changes of carriage of these agents in the absence of COVID-19 infection.
Conclusion
The COVID-19 pandemic and related containment policies have resulted in a decrease in invasive bacterial infections through a decrease in the circulation and transmission of these bacteria. The decrease in transmission is also associated with a decrease in viral infections, such as influenza. However, the presence of bacterial carriage of the three studied agents seems to be associated increase the need for oxygen therapy and intubation. The COVID-19 pandemic has shaken up bacterial and viral ecology and epidemiology. There is concern that viral and bacterial infections will re-emerge after the non-pharmaceutical control measures will be lifted. There is a clear interest in monitoring the post-pandemic evolution of the epidemiology of invasive bacterial diseases, especially those preventable by vaccination.
Declarations
Ethics approval and consent to participate
This study was approved by the Ethics Committee of [the Comité d’Ethique pour la Recherche sur Données Existantes et/ou hors loi Jardé du Centre Hospitalier Universitaire de Rouen] for Research on Existing Data of the C.H.U of Rouen. The need for informed consent was waived by the ethics committee/Institutional Review Board of [the Comité d’Ethique pour la Recherche sur Données Existantes et/ou hors loi Jardé du Centre Hospitalier Universitaire de Rouen] (France), because of the retrospective nature of the study. The study was performed in compliance with to the Declaration of Helsinki dated October 2013.
Consent for publication
Not applicable.
Availability of data and materials
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
Competing interests
No competing interests
Funding
Rouen University Hospital; Paris Institute Pasteur
Authors' contributions
Johnny Michel performed most of the laboratory manipulations with the help of Maria-Alexandra Stoica, Myriam Aouiti-Trabelsi, Fabienne De-Oliveira and Eva Hong. All authors have reviewed the manuscript in its entirety. Jean-Christophe Plantier and Muhamed-Kheir Taha are the principal directors of the project.
Acknowledgements
Not applicable.
References
- Zhu X, Ge Y, Wu T, et al. Co-infection with respiratory pathogens among COVID-2019 cases. Virus Res 285 (2020): 198005.
- Lansbury L, Lim B, Baskaran V, et al. Co-infections in people with COVID-19: a systematic review and meta-analysis. J Infect 81 (2020): 266-275.
- Contou D, Claudinon A, Pajot O, et al. Bacterial and viral co-infections in patients with severe SARS-CoV-2 pneumonia admitted to a French ICU. Ann Intensive Care 10 (2020): 119.
- Hoque MN, Akter S, Mishu ID, et al. Microbial co-infections in COVID-19: Associated microbiota and underlying mechanisms of pathogenesis. Microb Pathog 156 (2021): 104941.
- Smith AP, Williams EP, Plunkett TR, et al. Time-Dependent Increase in Susceptibility and Severity of Secondary Bacterial Infection during SARS-CoV-2 Infection. BioRxiv Prepr Serv Biol (2022).
- Ruan Q, Yang K, Wang W, et al. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med (2020):1-3.
- Garcia-Vidal C, Sanjuan G, Moreno-García E, et al. Incidence of co-infections and superinfections in hospitalized patients with COVID-19: a retrospective cohort study. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis 27 (2021): 83-88.
- Brundage JF. Interactions between influenza and bacterial respiratory pathogens: implications for pandemic preparedness. Lancet Infect Dis 6 (2006): 303-312.
- Völk S, P firrmann M, Koedel U, et al. Decline in the number of patients with meningitis in German hospitals during the COVID-19 pandemic. J Neurol (2022).
- Brueggemann AB, Jansen van Rensburg MJ, Shaw D, et al. Changes in the incidence of invasive disease due to Streptococcus pneumoniae, Haemophilus influenzae, and Neisseria meningitidis during the COVID-19 pandemic in 26 countries and territories in the Invasive Respiratory Infection Surveillance Initiative: a prospective analysis of surveillance data. Lancet Digit Health 3 (2021): e360-370.
- Taha MK, Deghmane AE. Impact of COVID-19 pandemic and the lockdown on invasive meningococcal disease. BMC Res Notes 13 (2020): 399.
- Danino D, Ben-Shimol S, van der Beek BA, et al. Decline in Pneumococcal Disease in Young Children During the Coronavirus Disease 2019 (COVID-19) Pandemic in Israel Associated with Suppression of Seasonal Respiratory Viruses, Despite Persistent Pneumococcal Carriage: A Prospective Cohort Study. Clin Infect Dis (2021): ciab1014.
- Camilli R, Vescio MF, Giufrè M, et al. Carriage of Haemophilus influenzae is associated with pneumococcal vaccination in Italian children. Vaccine 33 (2015): 4559-4564.
- Christensen H, May M, Bowen L, et al. Meningococcal carriage by age: a systematic review and meta-analysis. Lancet Infect Dis 10 (2010): 853-861.
- Deghmane AE, Hong E, Chehboub S, et al. High diversity of invasive Haemophilus influenzae isolates in France and the emergence of resistance to third generation cephalosporins by alteration of ftsI gene. J Infect 79 (2019): 7-14.
- Månsson V, Skaare D, Riesbeck K, et al. The spread and clinical impact of ST14CC-PBP3 type IIb/A, a clonal group of non-typeable Haemophilus influenzae with chromosomally mediated β-lactam resistance-a prospective observational study. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis 23 (2017): 209.e1-209.e7.
- Laboratory methods for the diagnosis of meningitis caused by Neisseria meningitidis, Streptococcus pneumoniae, and Haemophilus influenzae [Internet]. [cité 3 oct 2022]. Disponible sur: https://www.who.int/publications-detail-redirect/laboratory-methods-for-the-diagnosis-of-meningitis-caused-by-neisseria-meningitidis-streptococcus-pneumoniae-and-haemophilus-influenzae
- Real-time RT-PCR Protocol for the Detection of Avian Influenza A(H7N9) Virus [Internet]. [cité 3 oct 2022]. Disponible sur: https://www.who.int/publications/m/item/real-time-rt-pcr-protocol-for-the-detection-of-avian-influenza-a(h7n9)-virus
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23 (2006): 254-267.
- Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res 3 (2018): 124.
- Cho JH, Jung MY, Kim JK, et al. Detection of Bacteria in Normal Adult Nasal Cavity Based on Polymerase Chain Reaction–denaturing Gradient Gel Electrophoresis. Am J Rhinol Allergy 25 (2011): e18-22.
- Rawlings BA, Higgins TS, Han JK. Bacterial pathogens in the nasopharynx, nasal cavity, and osteomeatal complex during wellness and viral infection. Am J Rhinol Allergy 27 (2013): 39-42.
- Mackenzie GA, Leach AJ, Carapetis JR, et al. Epidemiology of nasopharyngeal carriage of respiratory bacterial pathogens in children and adults: cross-sectional surveys in a population with high rates of pneumococcal disease. BMC Infect Dis 10 (2010): 304.
- Amin-Chowdhury Z, Aiano F, Mensah A, et al. Impact of the Coronavirus Disease 2019 (COVID-19) Pandemic on Invasive Pneumococcal Disease and Risk of Pneumococcal Coinfection with Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): Prospective National Cohort Study, England. Clin Infect Dis (2021): e65-75.
- Peradotto M, Bondi A, Lombardi D, et al. The impact of COVID-19 pandemic control on vaccine-preventable invasive bacterial diseases in Piedmont (Italy). Infection (2022).
- Choi YH, Miller E. Impact of COVID-19 social distancing measures on future incidence of invasive pneumococcal disease in England and Wales: a mathematical modelling study. BMJ Open 11 (2021): e045380.
- Taine M, Offredo L, Drouin J, et al. Mandatory Infant Vaccinations in France During the COVID-19 Pandemic in 2020. Front Pediatr 9 (2021):666848.
- McDonald HI, Tessier E, White JM, et al. Early impact of the coronavirus disease (COVID-19) pandemic and physical distancing measures on routine childhood vaccinations in England, January to April 2020. Euro Surveill Bull Eur Sur Mal Transm Eur Commun Dis Bull 25 (2020).
- Masson E. Pediatric Infectious Disease Group (GPIP) position paper on the immune debt of the COVID-19 pandemic in childhood, how can we fill the immunity gap? [Internet]. EM-Consulte (2022).
- Clark S, Campbell H, Mensah AA, et al. An Increase in Group B Invasive Meningococcal Disease Among Adolescents and Young Adults in England Following Easing of COVID-19 Containment Measures (2022).
- Deghmane AE, Taha MK. Changes in Invasive Neisseria meningitidis and Haemophilus influenzae Infections in France during the COVID-19 Pandemic. Microorganisms 10 (2022): 907.


 Impact Factor: * 5.3
Impact Factor: * 5.3 Acceptance Rate: 75.63%
Acceptance Rate: 75.63%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 