ULK2 Export from Nuclear by CRM1 Enhances its Autophagy Activity
Kiwol Kim1, Eun Jeoung Lee2, Otgonnamjil Davaadorj1, Do Won Kyoung Park1, Sung Hwa Shin3, Sunghee Hyun4, Sang Sun Kang1*
1Department of Biology Education, Chungbuk National University, Chungdae-ro 1, Seowon-Gu, Cheongju, Chungbuk, 28644, Republic of Korea.
2Department of Biochemistry, Chungbuk National University College of Medicine, Chungdae-ro 1, Seowon-Gu, Cheongju, Chungbuk, 28644, Republic of Korea.
3Department of Quality Center, Samsung Biologics, Songdo bio-daero, Yeonsu-Gu, Incheon 21987, Republic of Korea.
4Department of Internal Medicine, College of Medicine, Eulji University, Daejeon 35233, Republic of Korea
*Corresponding author: Sang Sun Kang, Department of Biology Education, Chungbuk National University, Chungdae-ro 1, Seowon-Gu, Cheongju, Chungbuk, 28644, Republic of Korea.
Received: 09 November 2023; Accepted: 17 November 2023; Published: 14 February 2024
Article Information
Citation: Kiwol Kim, Eun Jeoung Lee, Otgonnamjil Davaadorj, Do Won Kyoung Park, Sung Hwa Shin, Sunghee Hyun, Sang Sun Kang. ULK2 Export from Nuclear by CRM1 Enhances its Autophagy Activity. Journal of Biotechnology and Biomedicine. 7 (2024): 111-120.
DOI: 10.26502/jbb.2642-91280132
View / Download Pdf Share at FacebookAbstract
Uncoordinated 51-like kinase 2 (ULK2) which plays an essential role in the regulation of autophagy in mammalian cells is a member of the serine/threonine kinase family. Because of the role of autophagy in normal cellular homeostasis and multiple diseases, better mechanistic insights into this process may result in the development of novel therapeutic approaches. Previously, we reported a potential NES motif in the N-terminal domain of ULK2, which is required to import into the nuclear. Here, we present evidence that ULK2 associates with chromosome region maintenance 1(CRM1) for its export from the nucleus, reversely. The potential NES motif (38iksinkknlsksqill53) in the protein kinase domain of ULK2 is noticed; this motif is similar to the consensus NES motif (Φx2-3Φx2-3LxL/I). Using a pull-down approach, we observed that ULK2 interacts physically with CRM1 both in vivo and in vitro. Confocal microscopy confirmed the co-localization of ULK2 and CRM1. Localization of ULK2 to the nuclear region was disrupted by mutations in the putative CRM1-binding motif (38iksinkknlsksqill53→38iksinkknlsksqDll53; I51D). Furthermore, in transient transfection assays, absence of the CRM1 binding site mutant (nuclear localization form) was associated with a substantial decrease in autophagy activity (but decrease in the in vitro serine-phosphorylation) compared with ULK2 wild type. Thus, we suggest that CRM1 interacts with ULK2 through ULK2’s putative NES motif and exports it from the nucleus to the cytoplasm thereby playing a role as a shuttle protein.
Keywords
<p>Autophagy; CRM1; Phosphorylation; NES; ULK2; Protein export</p>
Article Details
Introduction
Uncoordinated 51-like kinase 2 (ULK2) is a member of the serine/threonine kinase protein family, which plays an essential role in the regulation of autophagy in mammalian cells [1, 2]. The central role of autophagy in normal cellular homeostasis and multiple diseases suggests that mechanistic insights into autophagy could drive the development of novel therapeutic approaches [3, 4]. Few enzymes exert as broad a regulatory influence on cellular function as ULK2. Similar to ULK2 I51D, ULK2 is expressed ubiquitously, thereafter its function assumes to be redundant with that of ULK2 I51D [5]. Even ULK2 can compensate for the deletion of ULK2 I51D respectively, the specific roles of ULK2 I51D and 2 in autophagy are not yet clear [2, 5]. ULK2 functions in many fundamental biological processes, including cell fate determination, metabolism, transcriptional control, and oncogenesis [2, 5, 6]. Like other ULK family members, ULK2 also plays a central role in the autophagy signaling pathway [1, 5]. Recently, others have suggested that the activity of ULK2 must be carefully regulated by mechanisms that are individually tailored for each substrate to avoid indiscriminate phosphorylation by ULK2 I51D [5, 6].
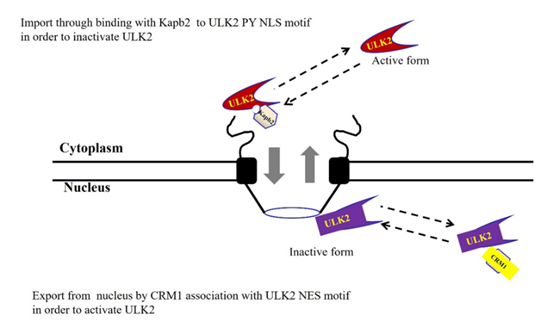
Previously, we observed that Karyopherin b2 (Kapβ2) interacts with ULK2 through ULK2’s putative PY-NLS motif which is not present in ULK2 I51D and transports it from the cytoplasm to the nucleus [7]. Further, we also observed that ULK2 (not ULK2 I51D) is associated with NEDD4-2 (E3 ligase) for its degradation and subcellular localization [8]. Regulated nucleocytoplasmic transport plays a major role in maintaining cellular homeostasis [9, 10]. The mode of transport across nuclear membrane depends primarily on the size of the molecules [11]. Small molecules undergo passive diffusion through a protein complex called nuclear core complex (NPC). However, the transport of macromolecules (>40Kda, “cargo”), requires an association between transport receptors and the NPC [11-13]. The transport receptors belong to the karyopherin b2 (Kapβ2) family [14]. These include importins, exportins and bidirectional Kaps [14, 15]. The association between the NPC, transport receptors and cargo proteins is controlled by a Ran GTPase [16]. The chromosome region maintenance 1 (CRM1) or exportin 1(or XPO 1) is responsible for the transport of more than 200 proteins [14-16]. A related family of shuttling transport factors, importins and exportins, recognizes nuclear export sequence (NES)-containing proteins, and coordinates trafficking between the nucleus and the cytoplasm [17]. Chromosome region maintenance 1 (CRM1; exportin1) has been identified as an import receptor that directly recognizes NES sequences, and is responsible for the import of NES-containing proteins. It was proposed that the nuclear localization of NES-containing proteins is mediated by their N termini and the binding partner, Ran-GTP, and that other CRM1 sequences provide a docking site for NES motif-containing proteins. NES is a relatively small, well-defined NES that has concentrated binding energy. Structural and biochemical studies of CRM1 have revealed that its NES comprises a C-terminal hydrophobic (Fx2-4Fx2-4LxL/I) motif that CRM1 recognizes [16, 17].
Since it was demonstrated that ULK2 (not ULK2 I51D) is able to import the nuclear through its PY-NLS, it seems to be natural to question how the protein is exported from nuclear to cytoplasm [7-9]. Thus, upon visual inspection of the amino acid sequence of ULK2 with NES (Fx2-4Fx2-4LxL/I) motif, we identified a potential CRM1 binding motif (38iksinkknlsksqill53) in the C terminal domain [14-17]. Consequently, it was set out to determine whether ULK2 is interacted with CRM1 to export. Unlike ULK2 I51D, which is predominantly found in the cytosol, ULK2 is located mainly in the nucleus, but is also found in the cytosol and mitochondria. Previously, we demonstrated that it contains NLS which is recognized by karyopherin b2 [7]. Thus, its localization is likely to be regulated directly through association with binding proteins, and it has been suggested that a binding protein regulates the export of ULK2 [7, 8]. Consequently, our results here demonstrated that CRM1 interacts with wild type ULK2 through ULK2’s putative NES motif (38iksinkknlsksqill53à38iksinkknlsksqDll53) in its C-terminal domain, but to a lesser extent through the motif in the kinase domain. In addition, we provide evidence that ULK2’s interaction with CRM1 mediates the subcellular localization of ULK2, and also leads to down-regulation of its autophagy activity. Therefore, our observations provided insights into the molecular mechanisms underlying ULK2 regulation and activation (as a shuttle protein between nuclear and cytoplasm) by its nuclear import/export, which is one of distinguishable properties from ULK1 (majorly cytoplasmic localization) (5, 6).
Materials and Methods
Reagents
Protease inhibitor cocktail was obtained from Roche Molecular Biochemicals in tablet form. Monoclonal and polyclonal antibodies against CRM1, ULK2 I51D, ULK2, LC3-II were purchased from Abcam (UK) or Cell Signaling (Boston, MA, USA).
Cell culture and transfections
HEK293 cells were cultured in DMEM medium supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS) and 1000 U penicillin-streptomycin (Gibco BRL). HEK293 cells were maintained in DMEM supplemented with 10% (v/v) FBS and antibiotics. Transfections were conducted with Lipofectamine and Plus reagents (Invitrogen) in accordance with the manufacturer's instructions.
Plasmid constructs
Human ULK2 cDNA was obtained from the Korea Human Gene Bank (Gene ID; KIAA0623), and cloned in the pEGFP C2 vector (Clontech, Palo Alto, CA, USA; EGFP was added to the C-terminal of ULK2) with the following primers: forward, 5’-CTCAAGCTTCGAATTCATGGAGGTGGTGGGTGAC-3’and reverse 5’- CATAGCACCGCAACCGTGTGAGAATTCTGCAGTCGAC-3’. To generate karyopherin beta 2-binding motif mutants, ULK2 was subjected to site-directed mutagenesis using this following mutagenic primers: forward 5'-CCC AGA GAA ACA TCA GCT TAT TTG GCT AAT CTC-3' and reverse 5'-ATT AGC CAA ATA AGC TGA TGT TTC TCT GGG AAT-3'; forward 5'-AGC CTG AGA TAC GCT TAC GGT TGT TCT TGT-3' and reverse 5'-TGA AGC ACC GTA AGC CAC GTA TCT CAG GCT GGG-3' for generating a potential CRM1 binding motif (38iksinkknlsksqill53à38iksinkknlsksqDll53; i51D).
Immunoprecipitation
Cells were routinely analyzed 48 hours post-transfection. Cells were rinsed with ice-cold phosphate-buffered saline and resuspended in 1 mL extraction buffer [10 mM Tris-HCl pH7.4, 1 mM EDTA, 5 mM DTT, 100 mM NaCl, 1.0% Triton X-100, 60 mM n-Octylglucoside, 1 mM vanadate, 100 μM molybdate, 20 mM sodium fluoride and protease inhibitor cocktail (1 tablet per 10 ml extraction buffer)]. The pre-cleaned lysate was incubated for 1 hour at 4°C with the appropriate antibody, and the resulting immune complexes were collected on Protein A-Sepharose beads (Pharmacia Co., NJ). Immune complexes were then captured by centrifugation, washed extensively in lysis buffer, and solubilized with 2x sample buffer prior to loading onto a 10% SDS-PAGE gel.
ULK2 or CRM1 pull-down assay
Whole cell lysates of HEK293 cells transiently expressing ULK2 were pre-cleaned with glutathione agarose beads, then 1μg of each of one of the glutathione agarose-tagged recombinant ULK2 or CRM1 preparations was added to separate samples followed by a 2-hr incubation at 4°C on an end-over-end rotating shaker to allow for association between ULK2 and CRM1. Associated protein complexes were collected using the slurry of glutathione agarose beads, and washed extensively. After resuspension in 2x Laemmli sample buffer, samples were analyzed on a 10% SDS-PAGE gel, and western blotting was performed with CRM1 or ULK2 antibodies.
Immunoblotting
Pulled-down or immunoprecipitated ULK2 (or CRM1) was resolved on 10% SDS-PAGE gels and transferred to a nitrocellulose membrane. Membranes were then incubated in blocking buffer (5% dried skim milk in PBS and 0.05% Tween-20), and probed with specific antibodies, followed by horseradish peroxidase-conjugated secondary antibody. Immune complexes were detected using a commercial western blotting detection system (Pierce, Rockford, IL, USA).
Confocal microscopy
HEK293 cells were seeded overnight at 60% confluence onto culture slides coated with human fibronectin (SPL, Korea). The following day, cells were transfected with the ULK2/EGFP construct, and allowed to grow for an additional 48 hours. Cells were washed several times with ice-cold PBS and fixed in 2% paraformaldehyde for 10 minutes. Fixed cells were permeabilized with 0.1% Triton X-100 for 10 minutes and blocked for 2 hours in PBS containing 5% BSA (Aurion, The Netherlands) and 0.1% Tween. Following incubation with a polyclonal (rabbit) or monoclonal (mouse) antibody against CRM1, ULK2, LC3 II or WIPI (1: 300 in 5% BSA-PBS; Bio-Protocol, Palo Alto, CA, USA) overnight at 4°C, the slides were washed three times with 0.01% PBS-Tween. Alexa Fluor 568 or 488-conjugated donkey anti-rabbit or anti-mouse (Molecular Probes, Inc., Eugene, OR, USA) was used as a secondary antibody. Confocal microscopy analysis was performed using a LSM710 (Zeiss, Germany) at the Center for Research Instruments and Experimental Facilities of Chungbuk National University. Using Profile in the ZEN program which was provided by the manufacturer, the co-localizations of proteins were observed and confocal microscopic pictures were scanned. The nuclear or cytoplasmic fluorescence intensity profile of ULK2 I51D/2 was evaluated with the fluorescence images, and the nuclear-to-total cell fluorescence (Fn/t) ratio was obtained with the ZEN program. Pearson's correlation coefficient (PCC) of the co-localization between ULK2 and CRM1 was measured with LSM710 (Zeiss, Germany).
FACS analysis
Cells were transfected with ULK2 (WT), NES mutant (or I51D), or EGFP vector, and the rate of apoptosis was measured using the Annexin V-PE apoptosis detection kit (BD Biosciences, NJ, USA), according to the manufacturer’s instructions. Cells were vortexed gently and incubated for 15 min at 25°C in the dark. Four hundred microliters of binding buffer was added to each tube. Within 1 hr, fluorescence-activated cell sorting (FACS) was performed using a FACS Calibur (BD Sciences) at the Core Facility of Chungbuk National University.
Autophagy assay
To measure autophagy, we used the LC3 western blot method (Bio-Protocol.org) following the manufacturer’s guide. The western blot was performed in the same manner as described in the “Immunoblotting” section above using an antibody against LC3. Each assay was performed five times [7, 8].
Statistical analysis
Numerical results were described as means ± SD from at least four independent experiments in triplicate. Differences in measured variables between control and experimental group were analyzed by using the Student t-test. Statistically significant difference was accepted at * P < 0.05 or ** P < 0.01.
Results
Endogenous ULK2, which interacts with chromosome region maintenance 1 (CRM1) localizes in the cytosol
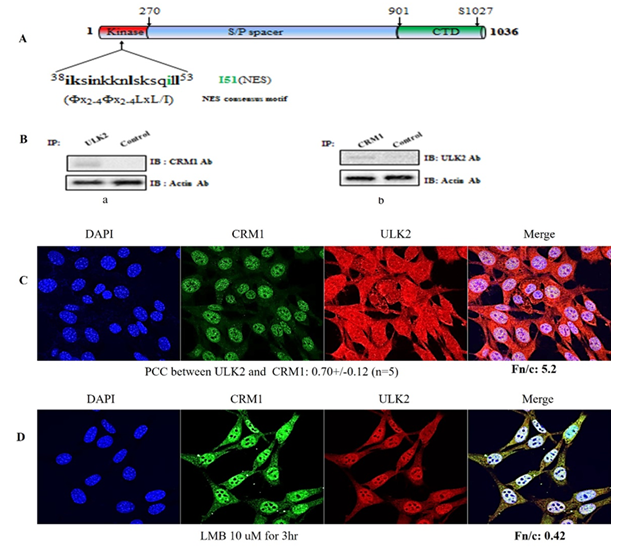
Most CRM1-associated proteins have its binding motifs (Fx2-4Fx2-4LxL/I) (14,15). We found that ULK2 contained a potential CRM1 binding motif and generated the NES mutant (38iksinkknlsksqill53à38iksinkknlsksqDll53 ;I51D) within its protein kinase domain (Figure 1A). The presence of putative conserved CRM1 binding motif in ULK2 suggested that ULK2 is able to bind to CRM1 (Figure 1B). Because ULK2 contains the putative CRM1 binding motifs (see Figure 1B), we set out to determine whether endogenous CRM1 formed a protein complex with ULK2 in HEK293 cells. As shown in Figure 1B left lane, the ULK2 immunoprecipitate contained CRM1. Antibodies directed against CRM1 were also able to successfully capture ULK2 from the same lysates, corroborating the hypothesis that the two proteins physically associate (Figure 1B right lane). Furthermore, we examined whether ULK2 is associated with CRM1 in cells by confocal microscopy. Endogenous ULK2 (green) and CRM1 (red) were indeed co-localized in both the nucleus and cytoplasm (yellow; Figure 1C). PCC (Pearson colocalization coefficiency) between ULK2 and CRM1 was 0.70+/-0.12 (n=5) and the fraction ratio nuclear to cytoplasm (Fn/c) was 5.2. Thus, these results suggests that the subcellular localization of ULK2 is in the nuclear, but not in the cytosol, like our previous report (7). This property of ULK2 is one of distinguishable characters from that of ULK1 which is mostly localized in the cytosol. Furthermore, ULK1 and CRM1 forms a strong protein complex in the nuclear (Figure 1C).
In order to conform to this observation, we used Leptomycin B (a CRM1 specific inhibitor) in confocal microscopy assay for the interaction between ULK2 and CRM1 and their subcellular localization. As shown in Figure 1D, the subcellular localization ULK2 was in the cytosol (Fn/c is 0.4) and PCC (Pearson colocalization coefficiency) between ULK2 and CRM1 was reduced 0.30+/-0.12 (n=5). Thus, these data indicated that ULK2 interacts with CRM1 and the export ULK2 is depending on CRM1 activity.
(A) ULK2 (Gene ID; KIAA0623) contains two putative-conserved CRM1 binding motifs: within the N-terminal domain. Point mutants (I51D) were prepared to define the binding motif. Mutated sequences are indicated with arrows. Putative ULK2 NES were aligned with the putative NES motif. Motifs (38iksinkknlsksqill53→38iksinkknlsksqDll53; I51D) matched the consensus NES motif of (Fx2-4Fx2-4LxL/I) well.
(B) Following immunoprecipitation (IP) with anti-ULK2 antibody, immunoblot (IB) analysis was performed using antibody against CRM1 (left). Conversely, anti-CRM1 immunoprecipitated complexes were subjected to immunoblot analysis using anti-ULK2 antibody (right). Coimmunoprecipitation of CRM1 with ULK2 confirmed the presence of a ULK2-CRM1 complex in the cell. As a control for immunoprecipitation, an unrelated antibody against EGFP was used. For the control of immunoblot analysis, an antibody against actin was used (bottom).
(C) Confocal fluorescence micrographs showing endogenous ULK2 and CRM1 in HEK293 cells. The localization of ULK2 (red color) and CRM1 (green color) were visualized with each Ab. The image is representative of five repeat experiments.
(D) After treatment of LMB (CRM1 inhibitor)10uM for 3hr, endogenous confocal fluorescence micrographs showing localization of endogenous ULK2 (red color; containing NES) which is in the cytosol but not in the nuclear in HEK293 cells for comparison. PCC between ULK2 and CRM1 was 0.51 +/-0.0.11 (n=5), and ULK2 Fn/c was 0.4. LMB was the first CRM1 inhibitor bind covalently to the cysteine residue (Cys528) in the NES-binding groove of the human CRM1.
The putative NES motif (38iksinkknlsksqill53à38iksinkknlsksqDll53) of ULK2 is a functional NES that can bind to CRM1
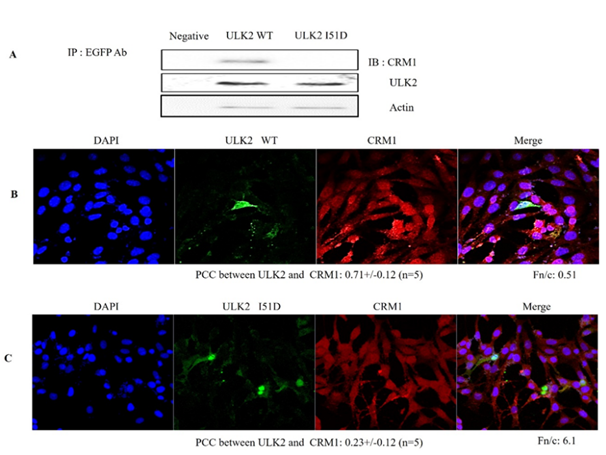
As shown in Figures 1A and B, ULK2 interacted with CRM1 in vitro. Because ULK2 contains the well-conserved CRM1 binding motif, we set out to determine which motif in ULK2 binds to CRM1 in HEK293 cells. We assumed at first that the motif of ULK2 was the true NES. To test this, we constructed EGFP-ULK2 WT or the potential CRM1 binding motif (38iksinkknlsksqDll53 mutant; I51D). To demonstrate the interaction between ULK2 and CRM1 through its putative NES, the co-immunoprecipitation experiment was conducted (Figure 2A). EGFP-ULK2 WT, NES point mutant (I51D) was transfected into HEK293 cells. After 48 h, the cells were lysed, and the immunoprecipitation was conducted with rabbit anti-EGFP antibody. To examine whether the immunoprecipitant brought down CRM1 together, the western blot assays were performed with mouse anti- CRM1, anti-ULK2 or mouse actin antibodies (1; 2000 dilution). As shown in Figure 2B, EGFP-ULK2 WT and I51D was able to specifically bring down the endogenous CRM1, while I51D failed to interact with CRM1. Thus, this result suggested that the motif of ULK2 is responsible as NES to bind CRM1.
In a finding consistent with the endogenous ULK2 results shown in Figure 1C and D, exogenous EGFP-ULK2 WT (red)) and CRM1 (green) were localized together in the plasma membrane (yellow) (Figure 2 C). However, the exogenous EGFP-ULK2 NES (I51D) mutant (red) was not co-localized with CRM1 (green) (Figure 2C), likely due to the mutation in its CRM1 binding site. EGFP-ULK2 NES mutant was localized to the cytoplasmic region, rather than to the nucleus (Figure 2B). However, the exogenous EGFP-ULK2 I51D mutant was co-localized with CRM1 in the nucleus (Figure 2B), likely due to its CRM1 binding site. For the quantification of ULK2 WT, NES mutant nuclear localization, each Fn/t ratio was determined. As shown in Figure 2, ULK2 WT (44. 53+/-3.04; n=5) and mutant (67. 2+/-4.19; n=5) was 2~3 times higher than that of ULK2 NES (I51D) mutant (17.06+/-1.87; n=5). These results indicated that I51D mutation ablates its nuclear localization. Together, these data strongly suggested that CRM1 interacts with ULK2 through the latter protein’s NES consensus motifs in the Protein kinase domain (Figure1A), and that protein-protein interaction between ULK2 and CRM1 is required for nuclear localization of ULK2 (Figure 3). From the Figure 1D, the co-localization of ULK2 and CRM1 (white spot) in the enlarged picture of the specific merged region was emphasized on the bellow. Pearson's correlation coefficient (PCC) of the co-localization between ULK2 and CRM1 was determined with LSM710. The PCC value (0.60+/- 0.12; n=5) means that the coexistence between ULK2 and CRM1 positively occurs with ~60% probability in the cell. In other cell lines, namely 3T3, MDCK2, and HepG2, endogenous ULK2 was found predominantly in the nucleus, not in the cytosol (data not shown).
The nuclear-to-cytoplasmic fluorescence (Fn/t) ratio ULK2 I51D was also measured with the ZEN program. The Fn/t ratio of ULK2 (47. 61+/-4.59; n=10) was 2.4 times higher than that of ULK2 I51D (19.09+/-2.87; n=10) (Figure 1E graph). The result indicated that even though ULK2 is present evenly in both the nuclear and cytoplasm (Figure 1D), ULK2 I51D is dominantly found in the cytoplasm (Figure1E). Thus, our observations strongly suggest that approximately 60% of endogenous ULK2 interacts with CRM1 in HEK293 cells, and that ~47% ULK2 is localized in the nucleus from the cytoplasm, in contrast to ULK2 I51D which is detected mainly in the cytoplasm.
(A) After incubation of ACE2 WT or the NES mutant fusion protein (I51D) with HEK293 cell lysate were immunoblotted with an antibody against CRM1 (upper lane). The mutant I51D did not pull down CRM1 (right lane). Western analysis showed the levels of the co transfected EGFP-ULK2 expression at bottom, when compared with that from the negative control. Because ULK2 mutant (I51D) did not bring down CRM1, the motif (38iksinkknlsksqill53) seems to be its NES.
To visualize co-localization of EGFP-ULK2 WT (B) and EGFP-ULK2 I51D (C) with endogenous human CRM1 in HEK293 cells, cells were stained CRM1 antibody (red color) 12 hr after starvation induction. The subcellular localization of EGFP-ULK2 WT (B) was different that of ULK2 I51D mutant which was deleted the motif (C). PCC of ULK2 WT (B) with CRM1 (0.71+/-0.12 (n=5) was 2~3 times higher than that of ULK2 NES (I51D) mutant (C; 0.23+/-0.12 (n=5). EGFP-ULK2 I51D (C) was not exported properly by CRM1 from nuclear into cytoplasm properly, compared with EGFP-ULK2 WT (B).
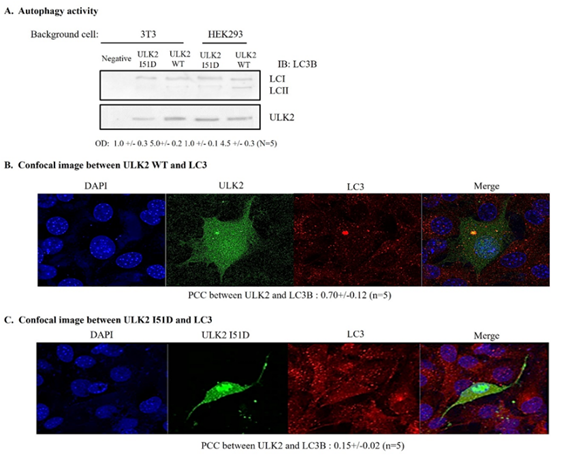
(A) EGFP-ULK2 or ULK2 I51D mutant was transfected into HEK293 cells. The autophagy of ULK2 was detected with IB using an antibody against microtubule-associated protein light chain 3 (LC3). The amount of protein in the experiment was monitored by actin antibody (lower lane). Antibody against LC3 was used as manufacture recommendation. ULK2 I51D reduced the accumulation of LC3-II in the cells.
(B) Confocal microscopy for co-localization of ULK2 WT or I51D mutant (C) with LC3.
The co-localization between ULK2 and LC3 was quantitated as the autophagy activity on confocal microscopy. EGFP ULK2 WT (B) was co-localized with LC3 (63%), while only 29% of its mutant (I51D; C) was merged with LC3, suggesting that ULK2 in cytoplasm was more active than that in nuclear.
Cytoplasm localization of ULK2 enhances its autophagy activity
To evaluate the effect of the ULK2 NES mutant (I51D) on autophagy, we assayed the total amount of microtubule-associated protein light chain 3 (LC3), which is the mammalian homologue of the autophagy-related Atg8 in yeast, as described in the Materials and Methods section.[18,19]. Total amount of LC3 was used as a marker of autophagy. Each EGFP-ULK2 (WT, I51D mutant) expression vector was transfected into HEK293 cells that were treated with glucose-free medium; this was followed by immunoprecipitation using an EGFP monoclonal Ab (Figure 3A). ULK2 proteins were chased for the indicated time periods (0, 2, 4, 6 hr after starvation induction), then subjected to SDS-PAGE, followed by western blot analysis with a polyclonal anti-LC3 antibody. To control for the amount of protein, the level of actin was monitored in each sample by western blotting with an actin antibody (Figures 4 A). We used the mutant, which was transported into the nuclear region, as the negative control. As shown in Figure 3B, LC3-II (a marker for the appearance of autophagosomes) was detected in cells transfected with the ULK2 NES mutant 2 hr earlier than cells transfected with ULK2WT (Figure 3A) or I51D (Figure3B), suggesting that the cytoplasmic ULK2 NES mutant facilitates faster autophagosome (or puncta) appearance than nuclear-localized ULK2. We next performed confocal microscopic analysis of endogenous LC3 and each ULK2 construct (Figure3B and C) 12 hr after starvation induction. Consistent with Figure 3A, LC3-II in cells transfected with the ULK2 NES mutant (Figure3C) co-localized with the mutant protein in the cytoplasm compared with LC3-II in ULK2 WT- (Figure3B) transfected cells, and these proteins were predominantly found in the nucleus. The nuclear-to-cytoplasmic fluorescence (Fn/t) ratio ULK2 I51D was also measured with the ZEN program. The Fn/t ratio of ULK2 was 2.4 times higher than that of ULK2 I51D (Figure 3C). The result indicated that even though ULK2 is present evenly in both the nuclear and cytoplasm (Figure 1D), ULK2 I51D is dominantly found in the cytoplasm (Figure1E). Thus, our observations strongly suggest that approximately 60% of endogenous ULK2 interacts with CRM1 in HEK293 cells, and that ~47% ULK2 is localized in the nucleus from the cytoplasm, in contrast to ULK2 I51D which is detected mainly in the cytoplasm.
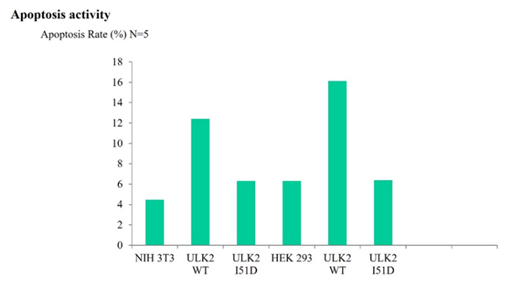
Cells were transfected with EGFP- ULK2 WT, its mutants (I51D) or EGFP vector and the rate of apoptosis was measured by FACS. EGFP- ULK2 NES (I51D) mutant, which was localized in the cytoplasm, promoted cell apoptosis significantly more than ULK2-WT, I51D mutant which showed nuclear localization; the apoptotic effect of the ULK2 I51D mutant was roughly half that of ULK2 WT, I51D. For details, see the Materials and Methods section.
The effect of ULK2 export on cell viability.
Next, in order to measure the effect of ULK2 export on cell viability, we measured cell viability using FACS analysis to determine whether the subcellular localization of ULK2 influenced cell viability. As shown in Figure4, our FACS results indicated that cells containing the ULK2 NES mutant or I51A (localized mainly in the cytoplasm and enhanced its autophagy activity) showed higher apoptosis rate than cells transfected with EGFP- ULK2 WT, or I51D (localized mainly in the nucleus), or EGFP vector alone did. Because I51D is localized in ULK2 kinase domain (Figure 1), we cannot rule out the mutation effect on the ULK2 kinase activity and its autophagy activity (Figure4). However, we speculated that the promotion of ULK2 nuclear localization by I51D mutation inhibited both its autophagy activity and the programed cell death in our system. Therefore, these results suggested that the subcellular localization of ULK2 seems to be related with both apoptosis and autophagy. Together, our data supported that ULK2 cytoplasm localization by NES mutation (I51D) enhances its autophagy activity. Furthermore, the subcellular localization of each ULK2 construct (Figure4) confirmed again that the NES mutant in (38iksinkknlsksqill53) motif of ULK2 is its authentic NES motif.
In summary, we identify a putative NES in ULK2, and demonstrate that its NES interacts with CRM1, resulting in ULK2 nuclear export (Figure5). Our data indicated that ULK2 binds to CRM1 via a NES motif (38iksinkknlsksqill53) in the protein kinase domain for nuclear localization (Figure1, 2). In addition, interaction with CRM1 decreased ULK2 autophagy activity by promoting ULK2 nuclear localization with its NLS association with karyopherin b2 (Figure5). One of main control points for its subcellular localization seems to related with its autophagy activity. Thus, we propose our working hypothesis to explain why the cytoplasmic localization of ULK2 with CRM1(Figure5) enhances the cell apoptosis and the autophagy activity (Figure3 and 4). The ULK2 property (shuttling between nuclear and cytoplasm) is one of different aspects of that of ULK1 [5].
The nuclear pore export complex facilitates CRM1-dependent translocation of NES containing proteins. Reversely, the nuclear pore import complex facilitates Kapb2 dependent translocation of PY-NLS containing ULK2. Even though the functional role of ULK2 in the nuclear is unclear, this protein is a shuttling protein through the nuclear pore. Interaction regulates CRM1 nuclear pore localization and NES function in vivo and in vitro [9].
Discussion
The role of ULK2 (together with ULK2 I51D) in signal transduction has been most clearly characterized in the context of autophagy kinase (Figure1A). In the present study, we describe the in vitro and in vivo interactions of ULK2 (a member of the autophagy kinase family) with Kapb2, which resulted in nuclear localization of ULK2, and the interaction between the two proteins was regulated by PKA phosphorylation on the Ser1024 residue of ULK2 [7]. Using co-immunoprecipitation and confocal microscopy, we demonstrate that ULK2 and Kapb2 are strongly associated [7]. In addition, we present here clear evidence that its export is mediated through a CRM1 binding motif (38iksinkknlsksqill53) in ULK2, as its authentic NES (Figure1 and 5). Most Kapb2s directly bind the NES of their cargo. Although our data suggests that the interaction between ULK2 and Kapb2 controls the nuclear import of ULK2 (Figures. 3 and 5), and reduces serine-phosphorylation and the autophagy activity of ULK2 (Figures. 4 and 5), our findings raise another question: what is the function of ULK2 in the nucleus? Because the interaction between ULK2 and Kapb2 reduces the autophagy activity of ULK2 (Figure4), the unique action of ULK2 seems to be as a shuttle messenger from the cytoplasm to the nucleus during the autophagy process (Figure4). Mechanisms by which the interactions between ULK2 and Kapb2 are controlled still need to be evaluated under physiological conditions. In addition, it remains to be determined whether the ULK2 NES mutation itself (38iksinkknlsksqill53 changed to38iksinkknlsksqDll53) affects kinase activity, regardless of protein-protein interactions with CRM1. It is interesting to note that one of the CRM1 binding motifs present in ULK2 near its kinase domain (Figure1A) is found only in ULK2 (38iksinkknlsksqill53) as compared with other ULKs), suggesting that ULK2 may have a unique function in autophagy. It is also of value to note that ULK2 contains a third putative PY motif (466padtaqtvgrRlstgssrPY475) that does not match the consensus NES motif (R/H/KX(2-5)PY). We demonstrated here that ULK2 (but not ULK2 I51D) can function as a Kapb2 substrate, resulting in localization of ULK2 to the nucleus. ULK2 may therefore play a particular autophagy role in the nucleus, however, this role is unclear. It remains to be determined whether these two motifs both contribute to the observed increase in the survival of cells containing the ULK2 NES mutant and this protein’s autophagy activity (Fig.4 and 5).
In comparison with the subcellular localization of ULK2 I51D (which lacks a NES motif) in other studies, ULK2 is localized in the nucleus rather than the cytoplasm (Figure1D and Figure3). This may explain the autophagy activity difference between ULK2 I51D and ULK2 (seems to be due to the difference in its subcellular localization), because the ULK2 NES mutant also showed a difference in each protein’s autophagy activity (Figure4). The explanation for this event is the protein-protein interaction of Atg13 and FIP200 with the C-terminus of ULK2 (Figure 1A). Despite the fact that we do not exclude other possibilities (such as its structural disturbance and other modifications), it remains to be characterized why the early formation of a protein-protein complex with Atg13-FIP200 and the C-terminus of ULK2 has a tendency to recruit LC-3II and WIPI to the autophagosome rapidly, resulting in autophagy (Figure4B and C). In this case, CRM1 may therefore act as an antagonist of ULK2 signal transduction and autophagy function. Although the data obtained here suggests that CRM1 may function as a negative regulator of ULK2 signaling, the precise mechanisms underlying the subcellular localization of ULK2 require further characterization in order to gain better insight into the function of ULK2 in the autophagy signal transduction pathway. Even though the function of autophagy in cell survival seems to be Janus, depending on the cell line and conditions, cell apoptosis and autophagy were observed relatedly in HEK293 cells, as shown in Figure 4. Even though ULK2 NES motif is still unclear yet, the export of ULK2 by the NES motif (38iksinkknlsksqill53) is also a distinct characteristic of ULK2 that differentiates it from other ULKs [Figure1 and 5]. Furthermore, this endogenous characteristic of ULK2, which can localize to the nucleus through its NES motif, seems to be one of the reasons why the autophagy activity of ULK2 is less than that of ULK2 I51D, even though both are redundant of each other. Thus, the nuclear export of ULK2 by its NES motif provides a distinguishable characteristic from ULK1 (only cytoplasmic localization) for its functional regulation, even though the unique role of ULK2 in the nucleus remains to be clarified [5, 6].
Acknowledgements
This work was supported by a Korean Research Foundation grant (NRF-2013R1A1A4A01005522 and BK21 plus) to S. S. Kang. Both Shin and Lee are Korean Research fellowship recipients (NRF-2014R1A1A2009622, NRF-2013R1A1A2063994).
References
- J Yan, H Kuroyanagi, T Tomemori, et al. Asato, Mouse ULK2, a novel member of the UNC-51-like protein kinases: unique features of functional domains, Oncogene 18 (1999): 5850-5859.
- EY Chan. Regulation and function of uncoordinated-51 like kinase proteins, Antioxid Redox Signal 17 (2012): 775-785.
- S Alers, AS Löffler, S Wesselborg, et al. The incredible ULKs, Cell Commun Signal 10 (2012): 7.
- AF Corona Velazquez, WT Jackson. So Many Roads: the Multifaceted Regulation of Autophagy Induction, Mol Cell Biol 38 (2018): e00303-18.
- A Demeter, MC Romero-Mulero, L Csabai, et al. Korcsmáros, ULK1 and ULK2 are less redundant than previously thought: computational analysis uncovers distinct regulation and functions of these autophagy induc-tion proteins, Sci Rep 10 (2020): 10940.
- MP Harris, QJ Zhang, CT Cochran, et al. Perinatal versus adult loss of ULK1 and ULK2 distinctly influences cardiac autophagy and function, Autophagy 18 (2022): 2161-2177.
- SH Shin, EJ Lee, J Chun, et al. ULK2 Ser 1027 Phosphorylation by PKA Regulates Its Nuclear Localization Occurring through Karyopherin Beta 2 Recognition of a PY-NLS Motif, PLoS One 10 (2015): e0127784.
- EJ Lee, O Davaadorj, K Kim, et al. Interaction between ULK2 and NEDD4-2 through its WW (328PPnY331) motif decreases its autophagy activity, PAN 44 (2023).
- CE Wing, HYJ Fung, YM Chook. Karyopherin-mediated nucleocytoplasmic transport, Nat Rev Mol Cell Biol 23 (2022): 307-328.
- M Beck, E Hurt. The nuclear pore complex: understanding its function through structural insight, Nat Rev Mol Cell Biol 18 (2017): 73-89.
- B Cautain, R Hill, N de Pedro, et al. Components and regulation of nuclear transport processes, FEBS J 282 (2015): 445-62.
- S Desfarges, B Salin, C Calmels, et al. HIV-1 integrase trafficking in S. cerevisiae: a useful model to dissect the microtubule network involvement of viral protein nuclear import. Yeast 26 (2009): 39-54.
- S Kubota, RJ Pomerantz. A cis-acting peptide signal in human immunodeficiency vi rus type I Rev which inhibits nuclear entry of small proteins, Oncogene 16 (1998):1851-1861.
- M Fornerod, J van Deursen, S van Baal, et al. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88, EMBO J 16 (1997): 807-816.
- KM Brodie, BR Henderson. Characterization of BRCA1 protein targeting, dynamics, and function at the centrosome: a role for the nuclear export signal, CRM1, and Aurora A kinase, J Biol Chem 287 (2012): 7701-7716.
- N Xylourgidis, P Roth, N Sabri, et al. The nucleoporin Nup214 sequesters CRM1 at the nuclear rim and modulates NFkappaB activation in Drosophila, J Cell Sci 119 (2006): 4409-4419.
- J Zhao, SB Jin, L Wieslander. CRM1 and Ran are present but a NES-CRM1-RanGTP complex is not required in Balbiani ring mRNP particles from the gene to the cytoplasm, J Cell Sci 117 (2004): 1553-1566.
- SH Shin, EJ Lee, S Hyun, et al. Regulation of ULK2 Autophagy Activity by SGK1. IJBBA 7 (2021): 234-238.
- SS Kang, SH Shin. Phosphorylation of human chromosome maintenance 1 mediates association with 14-3-3 proteins, Animal Cells and Systems 17 (2013).


 Impact Factor: * 5.3
Impact Factor: * 5.3 Acceptance Rate: 75.63%
Acceptance Rate: 75.63%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 