Accelerated Dynamic Time Warping on GPU for Selective Nanopore Sequencing
Harisankar Sadasivan1*, Daniel Stiffler2, Ajay Tirumala2, Johnny Israeli2, Satish Narayanasamy1
1Department of Computer Science and Engineering, University of Michigan Ann Arbor, MI 48109, USA
2NVIDIA Corporation, Santa Clara, CA 95051, USA
*Corresponding author: Harisankar Sadasivan, Department of Computer Science and Engineering, University of Michigan Ann Arbor, MI 48109, USA.
Received: 13 November 2023; Accepted: 22 November 2023; Published: 21 February 2024
Article Information
Citation: Harisankar Sadasivan, Daniel Stiffler, Ajay Tirumala, Johnny Israeli, Satish Narayanasamy. Accelerated Dynamic Time Warping on GPU for Selective Nanopore Sequencing. Journal of Biotechnology and Biomedicine. 7 (2024): 137-148.
DOI: 10.26502/jbb.2642-91280134
View / Download Pdf Share at FacebookAbstract
The design and supply of RT-PCR primers for accurate virus testing is a complex process. The MinION is a revolutionary portable nanopore DNA sequencer that may be used to sequence the whole genome of a target virus in a biological sample. Human samples have more than 99% of non-target host DNA and Read Until is a protocol that enables the MinION to selectively eject reads in real-time. However, the Min- ION does not have any in-built compute power to select non-target reads. SquiggleFilter is a prior work that identified the accuracy and throughput challenges in performing Read Until using the state-of-the-art solution and proposed a hardware-accelerated subsequence Dynamic Time Warping (sDTW) based programmable filter on an ASIC. However, SquiggleFilter does not work for genomes larger than 100Kb. We propose DTWax which optimizes SquiggleFilter’s sDTW algorithm onto the more commonly available GPUs. DTWax better uses tensor core pipes, 2X-SIMD FP16 computations and efficient data handling strategies using offline pre-processing, coalesced global memory loads, warp shuffles and shared memory buffering among other optimizations. DTWax enables Read Until and yields 1.92X sequencing speedup (improvement in sequencing time) and ∼3.64X compute speedup: costup (improvement in compute time normalized to cloud access cost) over a sequencing workflow that does not use Read Until.
Keywords
<p>Read Until; Selective sequencing; DTW; MinION; Metagenomics; Nanopore</p>
Article Details
Introduction
With SARS-CoV-2 evolving and adapting to its new environment [1] and becoming immune-evasive [2], there is a possible threat from a variant that can evade our current gold standard tests and fuel a surge in cases. Reverse Transcription Polymerase Chain reaction (RT-PCR) is the current gold standard [3] for SARS-CoV-2 diagnostic testing. Prior works have shown that RT-PCR requires the design and manufacture of custom PCR primers which is a complex, time-consuming, and error-prone process [4-6]. This limits the utility of RT-PCR in the early stages of a pandemic. Insufficient testing contributed to the uncontrolled spread of the virus early on. It took half a year after the first SARS-CoV-2 genome was sequenced in January 2021 for the number of daily tests worldwide to even cross 1 million [4]. Dunn and Sadasivan et al. [4] developed SquiggleFilter, a portable virus detector that could be re-programmed to speed up the sequencing of reads from a viral target of interest. SquiggleFilter is an ASIC, envisioned to work alongside Oxford Nanopore Technology’s (ONT) MinION MK1B (or simply the MinION), a recent-to-market portable DNA sequencer that does not have any compute built into it. However, SquiggleFilter can only be programmed with references of size less than 100Kb and it being an ASIC, is not easily scalable.
Additionally, GPUs are becoming a more common choice for accelerated computing on sequencers ONT sequencers GridION, PromethION, and MinION MK1C have GPUs built into them [7]. GPUs are also widely available in workplaces and on cloud platforms. While SquiggleFiter’s subsequence Dynamic Time Warping (sDTW) algorithm was optimized to work on an ASIC, we adapt and optimize it to work on the more common GPUs.
Background
Nanopore Sequencing
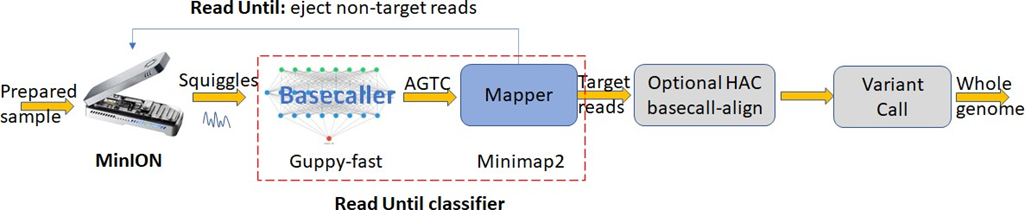
Long-read sequencing technology is increasingly becoming popular for rapid and accurate medical diagnosis [8,9] with lower adoption costs, end-to-end sequencing times, and improved portability and raw read accuracies. Long reads are particularly useful for applications that include structural variant calling and denovo assembly [8,10] as they, unlike short reads, can span highly repetitive regions in the genome. Oxford Nanopore Technology’s (ONT) MinION is a long-read DNA sequencer that is low-cost, real-time, portable, and can perform digital target enrichment using software instead of time-consuming wet-lab based methods [4]. While prior sequencing technology like Illumina relied on short accurate DNA reads, nanopore sequencers introduced long and noisy reads. ONT sequencers can produce very long reads to help span the highly repetitive regions in the genome. Nanopore senses the DNA molecule that passes through the pore by measuring the characteristic disruptions in electric current density. Decoding this noisy but characteristic signal (squiggle) helps us understand the DNA base (A, G, T, or C). MinION is an ideal candidate for viral detection because of many factors [4]. MinION is portable, low-cost, and capable of real-time DNA sequencing. Unlike RT-PCR tests, where one has to perform enrichment of target DNA in low-concentration specimens in the wet-lab, MinION lets us save time and cost by digitally checking for targets while sequencing. In real-time, MinION can be controlled to selectively sequence just the target DNA strands and eject the non-target strands by reversing the electrical potential across the pore.
Selective Sequencing
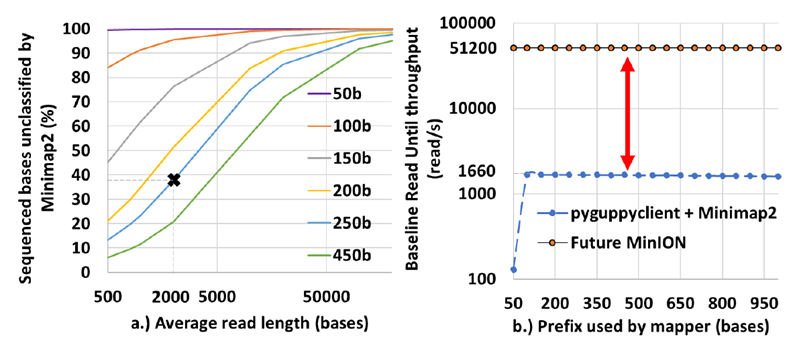
Most human samples have a very high fraction of non-target DNA (∼ 99.9% and most of it is human DNA) [11]. In order to save time and cost of sequencing, ONT has a feature called Selective Sequencing (Read-Until) [12] which lets us selectively sequence only the target DNA reads while ejecting the non-target. As the read is sequenced, real-time compute may be performed to classify the read as a target or not. Non-targets are ejected by reversing the voltage in the nanopore while targets are completely sequenced. The state-of-the-art selective sequencing pipeline [12,13] uses Guppy-fast (hereon referred to as Guppy) for basecalling and Minimap2 for classifying the reads [13] as shown in Figure 1. Guppy is a deep neural network-based software that converts the raw signal output of the MinION (noisy squiggles) to bases. However, Guppy has a two-fold performance problem. Prior works have demonstrated how Guppy is compute intensive [14-16] and does not have the required throughput even on a high-end edge GPU to handle the future throughput of the MinION [4] and SquiggleFilter [4] pointed out how the increasing throughput of the MinION amplifies this problem. We demonstrate the same problem in Figure 2b. Figure 2b also points to the fact that the GPU hardware is under-utilized when the prefix (query) lengths are smaller which we confirmed by profiling the software using NVIDIA Nsight Compute. Secondly, Guppy is unable to accurately basecall small chunks of data. ∼40% of the bases sequenced from a sample of average read length 2 Kbases is unclassified with prefix lengths of 250 bases as shown in Figure 2a because Guppy could not basecall these very short fragments accurately [16]. Please note that in this context, classification refers to mapping read prefixes to target (microbe) or non-target (host). Additionally, we trim the first 100 bases (or equivalent signal samples) to remove the adapters from all our analysis.
Figure 2: Guppy basecalls smaller signal chunks poorly leaving a high percent of sequenced bases unclassified by Minimap2. Guppy also has a throughput problem doing Read Until. (a) ∼40% of the bases sequenced are non-target in a 99.9: 0.1 non-target: target mix with an average read length of 2 Kbases and a Read Until read-prefix length of 250 bases. Read prefix lengths used for classification are color-coded. (b) Guppy followed by Minimap2 cannot match the throughput of a future MinION even on a high-end cloud instance that uses an A100 GPU for basecalling (prefix length used is 250 bases per second).
Hardware-accelerated SquiggleFilter [4] was proposed as a replacement for the Read Until classification using Guppy followed by Minimap2.
Prior Work
Since the MinION’s release in 2014, there has been a few attempts in improving the benefits of Read Until [4, 12-17], of which only SquiggleFilter [4] has the optimal combination of accuracy and throughput to keep up with a future MinION. The softwares performing Read Until can be broadly classified into two categories based on the inputs they operate on- signal space and basecalled space. Softwares operating in the signal space attempt to classify the input squiggle as a target or non-target while softwares operating in the basecalled space rely on the basecaller to transform raw squiggles to bases in real-time which is computationally expensive. We observe that the basecaller also basecalls smaller signal chunks poorly. The basecalled read prefixes may then be classified as a target or non-target. Readfish [13] and RUBRIC [12] classify basecalled reads to detect targets using mapping tools like Min- imap2. We show that ONT’s proprietary basecaller Guppy suffers from not being able to basecall smaller signal chunks correctly leading to ∼40% of the sequenced bases being unclassified by Minimap2. Addi- tionally, Guppy, a deep neural network, also suffers from low throughput on high-end GPUs and cannot meet the real-time compute requirements of a future MinION [4]. Three of the signal space-based methods rely on event segmentation- a pre-processing step where raw squiggles are segmented into events to detect positions in the signal where we are more likely to see a new base. The very first attempt at Read Until [18], UNCALLED [14], and Sigmap [15] use event segmentation as a pre-processing step. Loose et al. [18] uses sDTW on python to perform Read Until from events on a CPU. This yields sub-optimal performance. UNCALLED follows up with FM-index look-ups and seed clustering to find a target map. Although UNCALLED has a good mapping accuracy for smaller genomes, We observe that UNCALLED does not have the necessary throughput to match the compute requirements of a future MinION. Sigmap does seeding followed by Minimap2-style chaining on the seeds to identify a target map. But we observe that Sigmap needs a relatively long read prefix to identify a sufficient number of seeds and this turns out to be 4000 raw samples. Additionally, we observe that Sigmap has lower mapping accuracy than UNCALLED.
SquiggleNet [19] is a convolutional neural network-based software for classifying squiggles into target or non-target. However, SquiggleNet [19] is slower than guppy followed by Minimap2 and only achieves similar mapping accuracy to Guppy followed by Minimap2. SquiggleFilter [4] is a programmable ASIC that can match the throughput of a future MinION and yield optimal Read Until benefits. However, the initial cost of adoption is high as ASIC needs to be economically manufactured at scale and shipped in order to be deployed worldwide. GPUs on the other hand are already widely available at workplaces, shipped along with some of the sequencers, and also available on the cloud. DTWax is our proposed software which optimizes SquiggleFilter’s underlying sDTW algorithm on GPUs for Read Until. DTW has been parallelized in the past for various different applications on architectures including FPGAs [17, 20], Intel Xeon Phis [21], big data clusters [22], customized fabrics [23] and even GPUs [24,25]. HARU [17] is a recent work that implements SquiggleFilter’s algorithm on a budget-constrained FPGA. HARU cannot match the maximum throughput of the current MinION and is not a solution that can match the planned 100X throughput of the MinION. Crescent [26] is a recent closed-source implementation of SquiggleFilter’s algorithm directly on the GPU but ends up being 29.5X lower in throughput than DTWax possibly because of several reasons including not utilizing warp synchronized register shuffles for data sharing between threads and fewer number of cells computed per thread. cuDTW++ [27] is the best-performing prior work on GPU which accelerates DTW. However, cuDTW++ is ∼2.6X slower than DTWax and is built for database querying of very small queries and not for subsequence Dynamic Time Warping that is required to perform Read Until. Additionally, the normalization step is performed very inefficiently.
subsequence Dynamic Time Warping
sDTW is a two-dimensional dynamic programming algorithm tasked with finding the best map of the whole of the input query squiggle in the longer target reference. In sDTW’s output matrix computation, parallelism exists along the off-diagonal of the matrix and therefore, the computation happens in a wavefront parallel manner along this off-diagonal. Diagonals are processed one after the other. If the query is assumed to be along the vertical dimension of the matrix and the target reference along the horizontal dimension, the minimum score on the last row of the matrix will point to the best possible map of the query to the reference. This score may be compared to a threshold to figure out if the query is a target or not. The sDTW cost function is defined as follows:

Squiggle Filter
While traditional RT-PCR tests rely on complex custom primer design and time-consuming wet-lab processes for target enrichment, MinION can be controlled to selectively sequence only the target virus of interest using the Read Until feature. Utilizing the Read Until feature requires making real-time classifications during sequencing but the current MinION does not have any compute power. Dunn and Sadasivan et al. [4] demonstrated how basecalling is the bottleneck and constitutes ∼88-96% of Read Until assembly and how this problem was amplified with the projected 100X increase in ONT’s sequencing throughput. Their solution, SquiggleFilter [4], uses hardware accelerated subsequence Dynamic Time Warping (sDTW) to perform Read Until.
SquiggleFilter addresses the compute bottlenecks in portable virus detection and is designed to even handle the higher throughput of a future MinION. SquiggleFilter is programmable and offers better pandemic preparedness apart from saving time and cost of sequencing and compute. However, SquiggleFilter’s limited on-chip memory buffer only lets it test for viral genomes smaller than 100Kb. Additionally, SquiggleFilter uses a modified version of sDTW algorithm where the accuracy dip from various hardware-efficiency focussed optimizations are overcome with the match bonus [4]. Match bonus is a solution to a problem on the ASIC and performing this on the GPU can introduce branch divergence. We eliminate the match bonus, retain the assumption of reference deletions and optimize sDTW to run on GPUs.
Our contributions
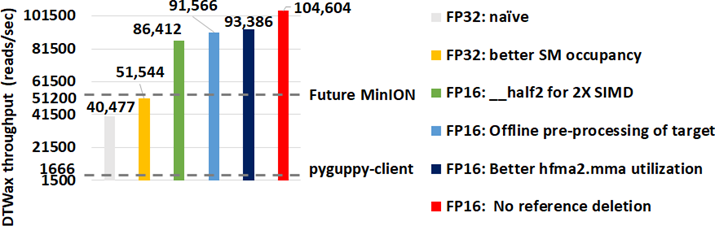
In this work. we present DTWax, a GPU-accelerated sDTW software for nanopore Read Until to save time and cost of nanopore sequencing and compute. We adapt SquiggleFilter ASIC [4]’s underlying sDTW algorithm to suit a GPU in order to overcome the limitation with reference lengths SquiggleFilter had. While sDTW was optimized for integer compute on the ASIC, we fine-tune sDTW for high throughput on the GPU. While SquiggleFilter uses integer arithmetic and Manhattan distances on the ASIC, we use floating point operations and Fused-Multiply-Add operations on the GPU. We also demonstrate how to utilize some of the GPU’s high throughput tensor core’s compute power for non-ML workloads.
As a first step, we speed up the online pre-processing step (normalization) on FP32 tensor cores using the batch normalization functionality from the CUDNN library traditionally used for machine learning workloads. DTWax is optimized to make use of the high throughput Fused-Multiply-Add instructions on the GPU. Further, we use FP16 and FP16 tensor core’s Matrix-Multiply-Accumulate (MMA) pipe for higher throughput for sDTW calculation. Using FP16 helps us process the forward and reverse strands, thereby extracting more parallelism to help improve the latency and throughput of classification. We also make use of offline pre-processing of reference squiggle index for coalesced loads, cudastreams for better GPU occupancy, intra-, and inter-read parallelism, register shuffles, and shared memory for low-overhead communication while processing the same query.
DTWax achieves ∼1.92X sequencing speedup and ∼3.64X compute speedup: costup from using nanopore Read Until for a future MinION (with 100X the current throughput) on an A100 compared to a workflow that does not use Read Until.
Methods
Offline pre-processing
ONT has published a k-mer current model [28] which provides a reference to map a 6-mer to an expected value of the current output from the MinION. We use this k-mer model to map the reference genome of the target virus to a noise-free FP16 squiggle reference. The squiggle reference will be of length (target_length - 6 + 1). We also pack two FP16 values (one from the forward strand and another from the reverse strand) into a half2 reference word (built-in CUDA datatype of two FP16 half-words). Further, we make use of the prior knowledge of the target reference to ensure coalesced global memory reads by re-ordering the target reference offline.
Online pre-processing: Normalization
The output squiggle of the MinION (query) is read from ONT’s proprietary FAST5 file format. The raw integer data is then scaled to pico-amperes (float32). The first few samples (1000) are trimmed to cut adapters and barcodes off. We re-purpose the CUDNN-Batchnorm to z-score normalize the 1-dimensional FP32 query current signal. CUDNN utilizes tensor cores and performs normalization at a very high throughout (∼6X the throughput of sDTW). The signal is then rounded off to FP16 and copied into a half2.
DTWax: architecture
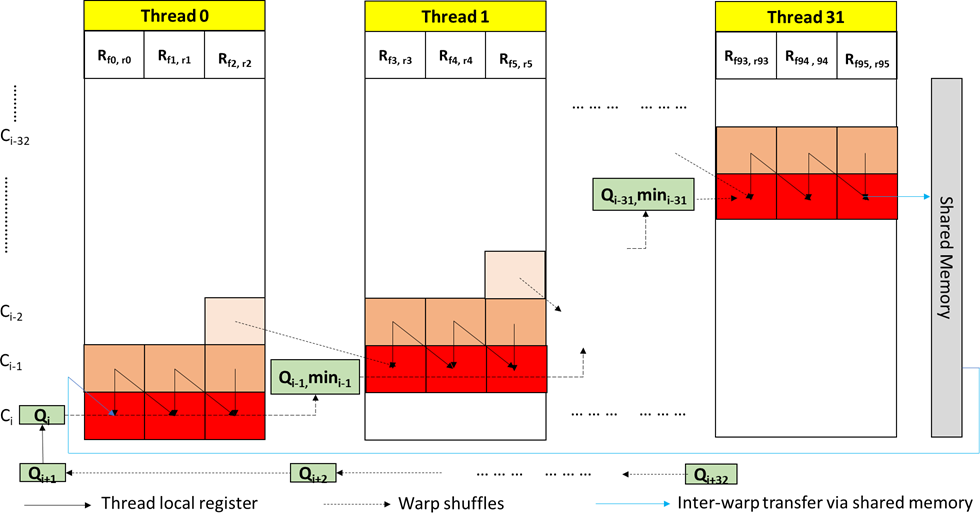
We adopt the segmented-sDTW architecture introduced in prior works [27] where each segment is a fixed number of cells in a row whose scores are computed by a thread. DTWax breaks down the processing of longer target references into multiple sub-matrices, each processing a fixed number of target bases. The reference length processed per sub-matrix is configurable and is set to 832 bases for optimal performance on an A100. Within a sub-matrix, each thread is responsible for processing a configurable but equal number of cells (cells per thread is called a segment). Wavefront parallelism exists along the off-diagonal segments in the sub-matrix as shown in Figure 3. Thread 0 is the first to finish its computation inside the sub-matrix while thread 31 is the last to finish. Target reference is loaded into registers (one FP16 reference sample each for forward and reverse strands into a single half2 datatype) from global memory using coalesced loads.
For intra-sub-matrix communication, we exploit warp shuffles for efficient register-to-register transfers within the same warp. This is an idea demonstrated by Schmidt et al [27] but not completely explored. Threads in a warp use warp shuffles to transfer the query sample, the minimum score of the segment, and the score of the last cell in the segment to the thread on its right. Instead of using a global reduction to find the final minimum score for DTWax, we use the efficient warp-shuffles to pass the minimum scores of the segments between threads. Inter-sub-matrix communication happens via shared memory transfers instead of relying on global memory. A thread block processing a read writes the last column of the sub-matrix into the shared memory and reads it back while calculating the consecutive sub-matrix for the same read.
Figure 3: Efficient intra- and inter-matrix communication in DTWax. Within a warp, each thread computes the FP16 vectorized scores for forward and reverse strands for a configurable segment number of cells in a wavefront parallel manner along the off-diagonal of the matrix. Warp shuffles are used for intra-warp communication (shown in dotted black) while shared memory is used as a buffer for inter-warp communication (shown in blue).
Intra- and inter-read parallelism
Using all the warps on an SM to process a single query would mean that the last warp remains idle and is ineligible for compute for an initial period of time. Therefore, we choose to process one read with a thread-block of only 32 threads. We have intra- and inter-read parallelism. Every query is processed by a thread block of 32 threads. Within a thread block, we have intra-read parallelism from 32 parallel threads each computing a segment of the sub-matrix. Across the GPU, we have inter-read parallelism as there as multiple concurrent blocks operating on different reads on any given Streaming Multi-processor (SM).
FP16 for 2X throughput
SquiggleFilter [4] has demonstrated that the information from the ONT sequencer may be captured using 8 bits. While the ASIC was custom-designed for integer arithmetic, GPUs are designed for high throughput floating point arithmetic. Among the floating point pipes available, we use the high throughput FP16 pipe on A100 (2X throughput compared to FP32) for DTWax. Computation with respect to the forward strand of the target reference happens on the first FP16 lane while the second FP16 lane computes with respect to the reverse strand. For example, Rf 0,r0 represents a FP16 vectorized forward and reverse reference signal points in Figure 3. Utilizing half2 FP16 pipes (FP16 vectorization) not only helps us to increase throughput but also improves the latency by 2X because we concurrently process both the forward and the reverse strand of the target with respect to the query in every cell of the sub-matrix.
Coalesced global memory access
The offline re-ordering of the target reference enables us to perform coalesced reads from global memory (as many loads as the length of one segment in a sub-matrix) before computation starts in the sub-matrix. The normalized target reference is an array of half2 datatype. This enables the vectorized processing of the input query signals on the high throughput FP16 pipe. Additionally, after the normalized query is read from the global memory in chunks of 32 half2 query samples using a coalesced load of 128B, it is then efficiently transferred between threads of a warp using warp shuffles. For example, in Figure 3, Rf 0,r0, Rf 3,r3, . . . , Rf 93,r93 would be read in one coalesced read.
Utilizing tensor core pipe
HFMA2.MMA pipe on the tensor core has one of the highest throughputs on A100. We re-formulate the addition in the cost function of DTWax to a Fused Multiply-Add operation in order to utilize the otherwise under-utilized HFMA2.MMA pipe. We are then able to better throttle the compute instructions between HFMA2.MMA and the remaining FP16 pipes instead of increasing the traffic on the FP16 pipe.
Assuming no reference deletion
Using the same assumption from SquiggeFilter [4] that viral strains have minimal reference deletions, we observe our accuracy of mapping using DTWax improves and our new cost function becomes simpler as we now only have to find a single minimum per cell instead of two minimums. This can be visualized in Figure 3 where is there no dependency on the immediate left neighbor’s score for score computation. Line 7 from Algorithm 1 is simplified to:
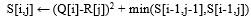
Optimizing occupancy and branch divergence
We ensured high SM utilization by finding the right balance between the number of resident warps on the SM and shared memory utilization. Further, we keep the GPU occupancy high by issuing concurrent asynchronous workloads to the GPU using cudastreams. Memory transfers to and from the GPU are overlapped with compute on the GPU. We reduce the branch divergence via partial loop unrolling. The first sub-matrix does not read from shared memory and the last sub-matrix does not write into shared memory. Unrolling the first and last sub-matrix computations of the query-target matrix helps improve performance.
Configurability and scalability
DTWax can be reprogrammed to test for any target reference of interest. Unlike some of the prior works [4, 27], DTWax can be reporgrammed to test for longer target references. Further, one may easily try and scale DTWax across multiple GPUs for higher throughput on longer or multiple target references.
Implementation
Experimental Setup
For all our GPU evaluations, we use Google Cloud Platform’s (GCP) a2-highgpu-1g instance with 40GB of memory and 85GB of host memory. The high throughput NVIDIA A100 GPU on this instance has the HFMA2.MMA pipe that we use for high throughput FP16 computations. To perform fair evaluations of prior works, our CPU baselines are configured to their best performance with hyper-threading enabled on GCP c2-standard-60 (30 Intel Cascade Lake cores and 240 GB of memory). Additionally, 10% higher GPU usage costs on GCP are factored in when benefits are measured.
pyguppy-client and DTWax use GPU while other softwares use CPU. We use UNCALLED v2.2, Sigmap v0.1, mappy (Python interface to Minimap2 v2.17) and pyguppy-client v0.1.0. We optimally configure the software for better mapping accuracy. We configure Sigmap for better event detection by setting “-min-num-anchors-output 2 -step-size 1”. We turn off minimizers in Minimap2 for better mapping accuracy by setting “-w=1 -k=15”.
As previously noted by Sadasivan et al. [16], benefits from Read Until can vary from run to run based on wetlab protocols (capture time and read length), quality of the flowcell and sample constitution. Therefore, it can be difficult to correctly measure the benefits of Read Until for multiple softwares across multiple live sequencing runs. To solve this, we utilize the analytical model from RawMap [16] to estimate sequencing time savings from using Read Until. We use the average capture time of 2 seconds observed by Sadasivan et al. [16] in wet-lab. The latency and throughput of above softwares used in Read Until are measured and plugged into this model as shown in the supplementary data. Additionally, the results presented from the analytical model also helps us to identify the optimal read-prefix length for Read Until classification and also to explore the benefits of Read Until on sequencing runs of different average read lengths. Although ONT’s Read Until API currently does not support read-prefix granularities (chunk sizes) lower than ∼200 bases (down from 400 bases) [13], it might be worthwhile to consider this as ONT’s support for short read length is increasingly growing.
We use NVIDIA Nsight toolkit for profiling software. NVIDIA Nsight Systems [29] is used to visualize concurrent CUDA events, and NVIDIA Nsight Compute [30] is used to profile GPU events.
We use public datasets used by SquiggleFilter [4]. The lambda phage DNA is sequenced on a MinION R9.4.1 flow cell following the Lambda Control protocol at the University of Michigan’s laboratory using the ONT Rapid Library Preparation Kit31. Human datasets (sequenced with MinION R9.4 and R9.4.1 flow cells) are obtained from ONT Open datasets [32] and the Nanopore Whole-Genome Sequencing Consortium [33]. In all our experiments and analysis, the first 100 bases (or equivalent samples) are trimmed to remove adapters.
Optimal GPU configurations
In order to optimize DTWax’s throughput on A100, we introduce more inter-read parallelism by fitting 32 blocks on each of the 108 Streaming Multiprocessors on the GPU. We process one read per thread block of size 32 threads. We observe that having multiple concurrent warps per read resident on the SM may not be beneficial as there may be a long latency in the last warp of a read getting valid inputs from the previous warp and starting to calculate useful values. Using Nsight Compute, we maximize the number of reference bases processed per thread (segment size) to 26 amounting to a total of 32x26 reference bases processed per thread block. In order to reduce global memory transactions, we use shared memory for inter-sub-matrix communication. A shared memory of size 500B is allocated per block to store the output of the last column of a sub-matrix calculated by the last thread in a thread block. This is read by the thread block when it processes the next set of 32x26 reference bases.
Incremental Optimizations
We modify Hund et al.’s software [27] to perform sDTW and observe that it is still slower than the future MinION. DTWax is a compute-bound kernel. We implement a series of optimizations to meet the throughput of the future MinION. Figure 4 gives the reader a better understanding of the relative benefits of some of the main optimizations that go into DTWax.
Results
We use two metrics to evaluate the benefits of using DTWax- sequencing speedup and compute speedup: costup. Sequencing speedup is defined as the speedup in the end-to-end sequencing time from using DTWax for Read Until over a conventional nanopore sequencing workflow that does not use Read Until (conventional workflow basecalls full length reads and maps them using Minimap2). Accessing an NVIDIA A100 GPU instance on the cloud is priced ∼10% higher than the CPU instance we use for benchmarking. Hence, we normalize the compute time savings from using DTWax for Read Until to the cost of the cloud GPU instance to estimate compute speedup: costup. Further, we also compare the F1-score of DTWax in making Read Until classifications. DTWax yields up to ∼1.92X sequencing
Figure 5: DTWax yields ∼1.92X sequencing speedup and ∼3.64X compute speedup: costup. (a) DTWax yields the best sequencing speedup of ∼1.92X over a conventional pipeline that does not use Read Until on reads of length 2000 bases. (b)DTWax yields the best compute speedup: costup of 3.64X. Prefix length used for classification is 250 bases for the best performance in both cases.
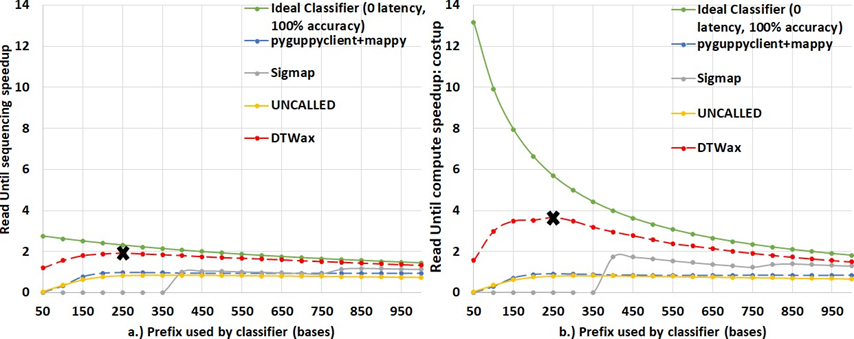
speedup and ∼3.64X compute speedup: costup with a future MinION of 100X throughput when compared to a sequencing workflow that does not use Read Until as shown in Figure 5. Additionally, we observe that using a prefix length of 250 bases yields the best benefits from using Read Until on a dataset of average read length 2 Kbases. One may also observe that savings from ont-pyguppy client degrades with increasing read-prefix lengths used for classification because ont-pyguppy cannot process streaming inputs and concatenate to outputs. Therefore, ont-pyguppy-client has to basecall the entire prefix again. Sigmap is unable to extract events from signals less than 400 bases. Please note that DTWax and pyguppy-client are GPU solutions and we label them using dotted lines on all the plots.
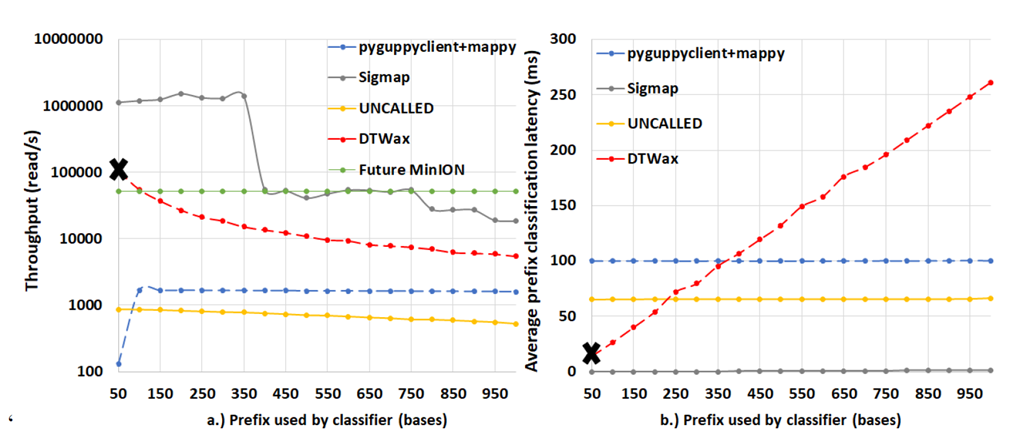
Figure 6: DTWax can handle more than 2X the throughput of a future MinION and has ∼7.18X lower latency than pyguppy-client followed by mappy. (a)Unlike pyguppy-client followed by mappy, DTWax operating at a granularity of 50 bases can handle twice the throughput of a future MinION (b)DTWax takes only ∼14 milliseconds to classify 50 bases and is ∼7.18X faster than pyguppy-client followed by mappy.
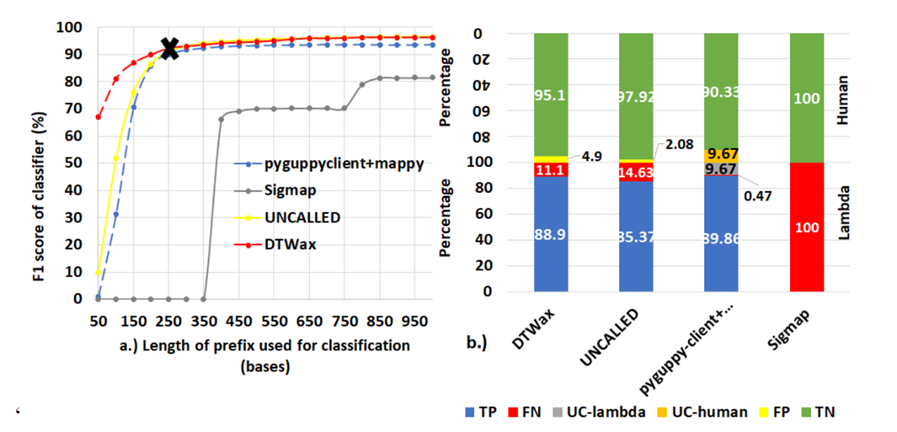
Figure 7: DTWax is the most accurate Read Until classifier. (a) DTWax has an F1-score of ∼92.24% and is the most accurate Read Until classifier using a prefix length of 250 bases. (b) DTWax is better at filtering non-target (human) reads out than pyguppy-client followed by mappy while being almost comparable in successfully retaining target reads
Figure 6 shows that DTWax can handle more than 2X the throughput of a future MinION and has ∼7.18X lower latency than pyguppy-client followed by mappy using a prefix length of 50 bases. Because of this, we operate DTWax at a processing granularity of 50 bases over a prefix length of 250 bases to make a Read Until classification decision. To explain the performance of pyguppy-client on longer prefixes, we profile pyguppy-client using NVIDIA nsight-compute and observe that the GPU occupancy is higher with longer prefix lengths resulting in better benefits from longer prefix lengths. UNCALLED seems to have a very high one-time fixed cost for path buffer management for storing output forest of trees and there is negligible cost with added prefix lengths. Although, Sigmap is the best in terms of throughput and latency, it cannot extract useful information from 250 bases and has lower F1 scores in classifying prefix lengths longer than 400 bases as shown in Figure 7.
Apart from the performance of the classifier, the correctness of classification also affects Read Until benefits. This is because a missed target read can mean sequencing another 1000 reads to collect a target read from a host: microbe sample mix of 99.9: 0.1. DTWax is the most accurate Read Until classifier as shown in Figure 7a. Figure 7a also shows that Sigmap cannot classify reads smaller than 400 bases. Figure 7b shows that DTWax is better at filtering non-target human reads and almost as good at retaining target reads when compared to pyguppy-client followed by mappy. We utilize the analytical model to explore the sequencing speedup and compute time savings from DTWax for higher average read lengths in Figure 8. Using the analytical model, we visualize the maximum benefit from Read Until that can come from an ideal classifier with zero latency and 100% accuracy of classification using the green line. DTWax is estimated to yield higher benefits on longer read lengths by the analytical model. However, it is worthwhile to note that this will only be possible with improvements in nanopore chemistry and pore design as currently long reads when ejected may tend to temporarily clog and block the sequencing pores.
Conclusions
SquiggleFilter is an ASIC that can be programmed to perform pathogen detection with the portable MinION sequencer. However, the ASIC cannot be programmed with references longer than 100 Kb. We adapt SquiggleFilter’s underlying sDTW algorithm to the more widely available GPUs to democratize Read Until. To make sDTW performant on the GPU, we do offline pre-processing of target reference to ensure coalesced loads from global memory, reduce branch divergence, and utilize FP16 vectorization and tensor core pipes. Further, we use warp-shuffles for efficient intra-sub-matrix communication and shared memory for low-latency inter-sub-matrix communication. Further, we assume no reference deletions to improve both the throughput and F1-score. We show that DTWax on an NVIDIA A100 GPU achieves ∼1.92X sequencing speedup and ∼3.64X compute speedup: costup over a sequencing workflow that does not use Read Until.
Acknowledgements
Development of DTWax was supported by NVIDIA Corporation and the University of Michigan Ann Arbor (via D. Dan and Betty Kahn foundation grants). Additionally, we would like to thank Google Cloud Platform for awarding us cloud research credits for the final evaluations of this research.
Author contributions statement
H.S. performed the analysis, design, implementation and evaluation of DTWax software apart from writing the manuscript. D.S. and A.T. recommended various performance optimizations and CUDA best practices for implementing DTWax based on DTWax’s profile. J.I. and S.N. led the collaborative effort and helped design the optimization strategy for DTWax. All authors reviewed the manuscript.
Data availability statement
Datasets used are published by SquiggleFilter here: :https://doi.org/10.5281/zenodo.5150973. One may download the dataset using the scrip provided by SquiggleFilter: https://github.com/ TimD1/SquiggleFilter/blob/master/setup.sh
Additional information
Software Availability
DTWax is open-sourced here: https://github.com/harisankarsadasivan/DTWax.
Competing interests
The authors declare that they have no competing interests.
References
- Amicone M, et al. Mutation rate of sars-cov-2 and emergence of mutators during experimental Evol. medicine, public health 10 (2022): 142-155.
- Willett BJ, et Sars-cov-2 omicron is an immune escape variant with an altered cell entry pathway. Nat microbiology 7 (2022): 1161-1179.
- Dramé M, et Should rt-pcr be considered a gold standard in the diagnosis of covid-19? J. medical virology 92 (2020): 2312.
- Dunn T, et al. Squigglefilter: An accelerator for portable virus detection. In MICRO-54: 54th Annual IEEE/ACM International Symposium on Microarchitecture (2021): 535-549.
- Park M, Won J, Choi BY, et al. Optimization of primer sets and detection protocols for sars-cov-2 of coronavirus disease 2019 (covid-19) using pcr and real-time pcr. & molecular medicine 52 (2020): 963-977.
- Patel Why the CDC Botched Its Coronavirus Testing. MIT Technology Review.
- Sadasivan H, et Accelerating minimap2 for accurate long read alignment on gpus. J Biotechnol Biomed 6 (2023): 13-23.
- Mantere T, Kersten S, Hoischen Long-read sequencing emerging in medical genetics. Front genetics 10 (2019): 426.
- Gorzynski JE, et Ultrarapid nanopore genome sequencing in a critical care setting. The New Engl Journal medicine (2022).
- Amarasinghe SL, et Opportunities and challenges in long-read sequencing data analysis. Genome biology 21 (2020): 1-16.
- Greninger AL, et Rapid metagenomic identification of viral pathogens in clinical samples by real-time nanopore sequencing analysis. Genome medicine 7 (2015): 1-13.
- Edwards HS, et Real-time selective sequencing with rubric: Read until with basecall and reference-informed criteria. Sci. Reports 9 (2019): 1-11.
- Payne A, et Readfish enables targeted nanopore sequencing of gigabase-sized genomes. Nat biotechnology 39 (2021): 442-450.
- Kovaka S, Fan Y, Ni B, et al. Targeted nanopore sequencing by real-time mapping of raw electrical signal with BioRxiv (2020).
- Zhang H, et Real-time mapping of nanopore raw signals. Bioinformatics 37 (2021): i477-i483.
- Sadasivan H, et al. Rapid real-time squiggle classification for read until using rawmap. Arch Clin Biomed Res 7 (2023): 45-47.
- Shih PJ, Saadat H, Parameswaran S, et al. Efficient real-time selective genome sequencing on resource-constrained (2022).
- Loose M, Malla S, Stout Real-time selective sequencing using nanopore technology. Nat. methods 13 (2016): 751.
- Bao Y, et al. Squigglenet: real-time, direct classification of nanopore signals. Genome biology 22 (2021): 1-16.
- Sart D, Mueen A, Najjar W, et al. Accelerating dynamic time warping subsequence search with gpus and fpgas. In 2010 IEEE International Conference on Data Mining (2010): 1001-1006.
- Kraeva Y, Zymbler M. Scalable algorithm for subsequence similarity search in very large time series data on cluster of phi In International Conference on Data Analytics and Management in Data Intensive Domains (2018): 149-164.
- Ziehn A, Charfuelan M, Hemsen H, et al. Time series similarity search for streaming data in distributed In EDBT/ICDT Workshops (2019).
- Xu X, et al. Accelerating dynamic time warping with memristor-based customized fabrics. IEEE Transactions on Des Integr Circuits Syst 37 (2017): 729-741.
- Hundt C, Schmidt B, Schömer E. Cuda-accelerated alignment of subsequences in streamed time series In 2014 43rd International Conference on Parallel Processing (2014): 10-19.
- Xiao L, Zheng Y, Tang W, et al. Parallelizing dynamic time warping algorithm using prefix computations on gpu. In 2013 IEEE 10th International Conference on High Performance Computing and Communications & 2013 IEEE International Conference on Embedded and Ubiquitous Computing (2013): 294-299.
- Li T, Li X, Li Y, et al. Crescent: A gpu-based targeted nanopore sequence selector. In 2022 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) (2022): 2357-2365.
- Schmidt B, Hundt cudtw++: Ultra-fast dynamic time warping on cuda-enabled gpus. In European Conference on Parallel Processing (2020): 597-612.
- Technologies, N. 6-mer model for r9.4 chemistry (2016). Oxford Nanopore Technologies (2016).
- Nsight NVIDIA https://developer.nvidia.com/nsight-systems.
- Nsight NVIDIA https://docs.nvidia.com/nsight-compute/NsightCompute/index.htmla.
- Rapid Library Preparation Kit (SQK-RAD004) (2021). Oxford Nanopore Technologies (2021).
- Technologies, N. Ont open datasets: Gm24385 dataset release (2020).
- Workman RE, et Nanopore native rna sequencing of a human poly (a) transcriptome. 459529 (2018).


 Impact Factor: * 5.3
Impact Factor: * 5.3 Acceptance Rate: 75.63%
Acceptance Rate: 75.63%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 