Renalase Identified by Machine Learning Methods as A Novel Independent Predictor of Mortality in Hospitalized Patients with COVID-19
Basmah Safdar1*, Matthew Sobiesk2, Dimitris Bertsimas2, Armin Nowroozpoor1,3, Yanhong Deng4, Gail D’Onofrio1, James Dziura1,4, Joe El-Khoury5, Xiaojia Guo6,7, Michael Simonov, R Andrew Taylor1, Melinda Wang7, Gary Desir6,7
1Department of Emergency Medicine, Yale School of Medicine, New Haven, Connecticut, United States
2Operations Research Center, Sloan, MIT, Cambridge, Massachusetts, United States
3Department of Emergency Medicine, Duke University School of Medicine, Durham, North Carolina, United States
4Yale Center for Analytics, Yale School of Medicine, New Haven, Connecticut, United States
5Department of Laboratory Medicine, Yale School of Medicine, New Haven, Connecticut, United States
6VACHS, West Haven, Connecticut, United States
7Department of Internal Medicine, Yale School of Medicine, New Haven, Connecticut, United States
*Corresponding author: Basmah Safdar, Department of Emergency Medicine, Yale School of Medicine, New Haven, Connecticut, United States.
Received: 12 January 2024; Accepted: 22 January 2024; Published: 12 March 2024
Article Information
Citation: Basmah Safdar, Matthew Sobiesk, Dimitris Bertsimas, Armin Nowroozpoor, Yanhong Deng, Gail D’Onofrio, James Dziura, Joe El- Khoury, Xiaojia Guo, Michael Simonov, R Andrew Taylor, Melinda Wang, Gary Desir. Renalase Identified by Machine Learning Methods as A Novel Independent Predictor of Mortality in Hospitalized Patients with COVID-19. Journal of Biotechnology and Biomedicine. 7 (2024): 175-185.
DOI: 10.26502/jbb.2642-91280139
View / Download Pdf Share at FacebookAbstract
Background: Low levels of renalase, a flavoprotein released by kidneys, has been linked with cytokine release syndrome and disease severity of viral infections. We sought to, 1) identify traditional and novel predictors of mortality for patients hospitalized with COVID-19 using traditional and machine learning methods; and 2) investigate whether renalase independently predicts mortality using these techniques.
Methods: In a retrospective cohort study, clinicopathologic data and blood samples were collected from COVID-19 patients hospitalized between March 1 and June 30, 2020. Patients were excluded if <18 years or opted out of research. Novel research markers – renalase, kidney injury molecule-1, interferon (a,d,i), interleukin (IL-1, IL6), and tumor necrosis factor were measured. The primary outcome was mortality within 180 days of index visit.
Results: Among 437 patients who provided 897 blood samples, mean age was 64 years (SD±17), 233 (53%) were males, and 48% were non-whites. Seventy-one patients (16%) died. Area under the curve (AUC) for mortality prediction was as follows: using logistic regression with a priori feature selection (AUC=0.72; CI 0.62, 0.82), logistic regression with backward feature selection (0.70; CI 0.55, 0.77), and XGBoost (0.87; CI 0.77, 0.93)]. PR-AUC and calibration plots also showed best performance with XGBoost model. Elevated BNP, advanced age, oxygen saturation deviation, and low renalase were the leading predictors of mortality in XGBoost. Renalase emerged as an independent predictor of mortality for COVID-19 across all statistical models.
Conclusion: Machine learning methods augment traditional statistical methods in identifying novel predictors of mortality such as renalase in patients with COVID-19.
Keywords
<p>COVID-19; Mortality; Biomarker; Inflammation; Cardiac; Renalase; Prediction; Machine learning</p>
Article Details
Abbreviations:
COVID-19: Coronavirus disease 2019; EHR: Electronic health records; CRP: C-reactive protein; IL-1: Interleukin 1; IL-6: Interleukin 6; TNFα: Tumor necrosis factor alpha; ARDS: Acute respiratory distress syndrome; ML: Machine learning; SARS-CoV-2: Severe acute respiratory syndrome coronavirus 2; RT-PCR: Reverse transcription-polymerase chain reaction; ELISA: Enzyme-linked immunosorbent assay; INF: Interferon; KIM-1: Kidney injury molecule 1; BNP: Beta natriuretic peptide; SD: Standard deviation; AUC: Area under the curve; PR-AUC: Precision recall area under the curve; RNLS: Renalase; ACE-2: Angiotensin-converting enzyme 2; RAAS: Renin-angiotensin-aldosterone; HIF 1-alpha: Hypoxia inducible factor; STAT3: Signal transducer and activator of transcription 3; MAPK: Mitogen activated protein kinase; AT1R: Angiotensin 1 receptor; NF-kb: Apoptosis nuclear factor
Introduction
COVID-19 has caused devastating morbidity and mortality. As of 2023, there were over 659 million reported cases and 6.6 million deaths [1]. Since the advent of the pandemic, clinicians and researchers have searched for predictors of mortality to help health systems better identify COVID-19 patients at high-risk, to guide treatment, and to efficiently allocate resources. Prior results have utilized data typically available within the electronic health records (EHR), such as clinical observations that outline a higher-risk phenotype (e.g., elderly male, obese, with coronary artery disease or hypoxia on presentation) and blood markers (such as cardiac markers, inflammatory markers, and coagulation markers). Given the novelty of COVID-19, the relative weight of these predictors seems to change with each cycle of the infection. Significant knowledge gaps remain, and there is a pressing need for further biomarker risk assessment that might uniquely contribute to pathogenesis, prognosis and treatment response. Several biomarker studies point to a unique immune response to COVID-19. Patients who die appear to have a pathophysiology characterized by an aggressively disordered inflammatory response distinct from those with milder symptoms. COVID-19 mortality is associated with increased hematological [e.g. leukocytes], inflammatory [e.g. ferritin, C-reactive protein (CRP), procalcitonin] markers, cytokines [e.g. interleukins (IL-1 and IL-6), and tumor necrosis factor (TNF-alpha) [2]. The immune response, however, appears to be much more muted in COVID-19 compared to disease states such as acute respiratory distress syndrome (ARDS) from sepsis or in cancer patients [3]. This observation questions the role of cytokine storm alone playing a primary role in the pathogenesis. Autopsy reports of COVID-19 patients indicate extensive vascular injury and consumption pathology (e.g., platelets), highlighting a need to look further for novel therapeutic targets. Changes in some markers, such as that of renal injury, appear to be transient [4].There is also scant information on markers that are depleted in COVID-19 or the dynamic course of these markers.
Renalase is one such peptide endogenously produced by the kidney, heart, and endothelium, with pro-survival properties such as reducing cytokine release in viral infections, including COVID-19 in mouse models (our unpublished data). We have previously shown low serum renalase to be associated with higher mortality in hospitalized patients with severe COVID-19 [5]. In a different cohort, renalase levels appear to rise in hospitalized patients with COVID-19 who survive and are ultimately discharged [6]. Renalase has also been linked to mitochondrial function and ATP production, suggesting that renalase is involved in the metabolic repair mechanisms of renal injury in mouse models [7]. However, it remains unclear how dynamic changes in endogenous renalase levels may influence survival in patients with severe COVID-19. To examine these issues, we leveraged traditional statistical approaches, newer machine learning (ML) models, [8] and a comprehensive prospective COVID-19 registry tracking the entire hospitalization, enhanced by testing of novel markers using serial serum samples. Traditional models of identifying predictors of mortality for COVID-19 have some predictable biases, are dependent on known literature, and tend not to perform as well when externally validated. ML approaches, while potentially less interpretable, have previously been demonstrated to offer performance advantages and generalizability over traditional methods, while also better factoring in variable interactions. For this study our goals were to, 1) identify and compare traditional and novel predictors of mortality for patients hospitalized with COVID-19 using traditional and ML techniques; and 2) assess if renalase independently predicts mortality using these methods.
Methods
Patient population
We conducted this study in hospitalized adult patients in a large, urban academic center between March 1 and June 30, 2020. All patients had confirmed severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by RT-PCR of nasopharyngeal swab samples. All specimens and imaging were collected as part of routine medical care. A standard clinical protocol for treatments was implemented during the study period. Exclusions included patients <18 years, those who opted out of research on admission, or had insufficient data. The protocol was approved by the Yale Institutional Review Board (HIC 2000027792, 2000028383 and 2000027690).
Clinical and laboratory data abstraction
We used Department of Medicine COVID Explorer (DOM-CovX), a cohort of patients hospitalized with COVID, to extract the clinical data from EPIC (electronic medical record system), including socio-demographics, comorbidities, vital signs, laboratory measurements, procedures, and disposition over the course of entire hospital stay [9]. Manual chart reviews were conducted to abstract admission date, presenting symptoms, smoking history, immunocompromised status, cardiopulmonary resuscitation and dates of intubation, death and last follow-up completed. Available serum or plasma samples were assayed from this cohort as follows: a) renalase levels were measured using denaturation (acid)-sensitive pool by ELISA; [10] and b) inflammatory markers, including IFNγ, IFNλ, IFNα, IL-1β, IL-6, and TNFα, as well as c) kidney injury molecule (KIM-1) were also measured using MSD V plex assays (Meso Scale Diagnostics, LLC, Rockville, MD), according to manufacturer’s instruction.
Variable definitions
Primary outcome was defined as death within 180 days of index visit. Hypoxia was defined as <90% oxygen saturation. We labeled cardiac markers as (high sensitivity troponin, beta natriuretic peptide [BNP]), markers of inflammation as (interleukin IL-6 and IL-1), and markers of thromboembolism as (D-dimer and platelets).
Statistical analysis
The data were presented as percentages for categorical variables and means and standard deviations (SD) for continuous variables. For univariate analyses, categorical variables between groups were compared using Fisher's exact test. Renalase trajectories (low to low, low to high, etc.,) were compared with other biomarker trajectories using the Chi-Square test.
Data Preprocessing
Categorical variables were one-hot encoded. Continuous variables with multiple values (e.g., oxygen saturation) were aggregated into maximum, minimum, and median values. Cutoffs between low and high values of biomarkers were determined by introspection of quantiles and clinical significance: BNP (100 ng/L), high sensitivity troponin (18 ng/ml), platelets (170 k/ul), IL-1 (0.02 pg/ml), IL-6 (8.66 pg/ml) and renalase (6000 ng/ml). Features with more than 30% of values missing were removed from the analysis (Supplement Table S5) [11]. To impute missing data, we utilized OptImpute, ML approach that has outperformed other related extant methods [12].
Model Development
A combination of traditional and newer ML methods was used. For traditional models, we used logistic regression and for newer methods, we used XGBoost. The latter is an aggregation of multiple decision tree models, which partition covariates in a hierarchical manner, dividing the input data points into disjoint sets [13]. XGBoost trains different trees against each other and then makes a final prediction by aggregating the results from all the different trees. In a study which compared 13 different algorithms on 165 bioinformatics to identify which algorithm performed the best in general, the authors found that gradient boosted trees outperformed all other algorithms significantly [14]. XGBoost model results can be exported to more easily interpretable SHAP values that allow readers to better understand the role of different covariates play in the model [15]. They are calculated by analyzing how much a feature impacts the final prediction of the model, and how the prediction itself is affected by the feature. This results in an ordered list of features, with information about whether higher values for a feature result in a positive or negative prediction.
A traditional logistic regression model and an XGBoost model were trained on all available features after applying exclusions due to missingness. An additional traditional logistic regression model was created using clinically relevant confounders that were selected a-priori based on univariate analysis findings, front line clinical experience and review of published data to serve as a baseline. These included age, sex, race, disease severity, BMI, smoking status, history of hypertension, chronic pulmonary disease, coronary artery disease, immunocompromised status, CRP, IL-6, renalase, creatinine, time from admission and time for initial symptom to blood sample [16].
Model Evaluation
The main metric used to assess the quality of the models was Area under the curve (AUC) and precision recall curve (PR-AUC). The AUC values of the model measure its ability to differentiate patients who would die of COVID-19 from patients who would not, where randomly guessing would result in an AUC of 0.5. Fifty percent of observations were randomly assigned to the training set, 20% to the validation set and 30% to the testing set. The models were fit on the training set and then the out-of-sample AUC value for the validation set was computed for different choices of parameters. The parameters of the model that yielded the highest AUC for the validation set were selected, and these parameters were used to train a model on the combined training and validation sets. The final out-of-sample AUC scores we present were calculated on the test data set. To generate confidence intervals for the AUCs, models were trained using 200 splits of the data, and mean values and confidence intervals were calculated empirically. Calibration plots were generated for example models for logistic regressions and XGBoost to analyze the different relationships between their predictions and the true outcomes. Backwards step regression was utilized in logistic regression models, using AIC for elimination. For XGBoost, we tuned the depth hyperparameter from 3 to 10, and the eta hyperparameter from 0.1 to 0.9. Logistic regression models were trained in R (v. 2014; http://www.R-project.org/), and XGBoost was trained in Python3 using the package created by the authors of the paper.
Results
Between March 2020 and June 30, 2020, 3450 patients with COVID-19 were admitted. Of these, 473 patients opted in for research, were >18 years and provided sufficient blood samples. Compared to the hospitalized patients not enrolled (n=2977), patients in our cohort were similar in age (mean age 63 vs. 63.8 years) and sex distribution (51% vs. 53% males) [5]. Additionally, 36 patients were excluded for having a missing date for index visit, negative follow-up time, or not having Covid-19, providing 437 patients for analysis.
|
Demographics |
Total (n=437) |
Survived (n=366) |
Died (n=71) |
|
Age; mean (SD) |
63.8 (17.0) |
61.8 (16.8) |
74.3 (14.6) |
|
Male; n (%) |
233 (53.3%) |
191 (52.2%) |
42 (59.2%) |
|
Hispanic; n (%) |
82 (18.8%) |
74 (20.2%) |
8 (11.3%) |
|
Race; n (%) |
|||
|
White |
228 (52.2%) |
182 (49.7%) |
46 (68.4%) |
|
Black |
132 (30.2%) |
114 (31.4%) |
18 (25.4%) |
|
Other |
77 (17.6%) |
70 (19.1%) |
7 (9.9%) |
|
Past Medical History |
|||
|
Hypertension; n (%) |
308 (70.5%) |
247 (67.5%) |
61 (85.9%) |
|
Diabetes; n (%) |
176 (40.3%) |
144 (39.3%) |
32 (45.1%) |
|
Hyperlipidemia; n (%) |
176 (40.3%) |
147 (40.2%) |
29 (40.8%) |
|
Myocardial Infarction; n (%) |
47 (10.8%) |
35 (9.6%) |
12 (16.9%) |
|
Congestive Heart Failure; n (%) |
101 (23.1%) |
72 (19.7%) |
29 (40.8%) |
|
Chronic Pulmonary Disease |
158 (36.2%) |
129 (35.2%) |
29 (40.8%) |
|
Chronic Kidney Disease; n (%) |
99 (22.7%) |
77 (21.0%) |
22 (31.0%) |
|
Immunocompromised; a n (%) |
72 (16.5%) |
57 (15.6%) |
15 (21.1%) |
|
Pregnancy; n (%) |
7 (1.6%) |
7 (1.9%) |
0 (0.0%) |
|
Smoking; n (%) |
182 (41.6%) |
147 (40.2%) |
35 (49.3%) |
|
Symptoms at Presentation |
|||
|
Chest pain; n (%) |
54 (12.4%) |
49 (13.4%) |
5 (7.0%) |
|
Cough; n (%) |
293 (67.0%) |
255 (69.7%) |
38 (53.5%) |
|
Fever; n (%) |
321 (73.5%) |
274 (74.9%) |
47 (66.2%) |
|
Dyspnea; n (%) |
264 (60.4%) |
226 (61.7%) |
38 (53.5%) |
|
Gastrointestinal symptoms; n (%) |
133 (30.4%) |
122 (33.3%) |
11 (15.5%) |
|
Admission |
|||
|
BMI; mean (SD) |
30.1 (7.6) |
30.5 (7.6) |
27.7 (6.8) |
|
Pulse; mean (SD) |
82.5 (16.1) |
81.5 (15.3) |
87.5 (18.9) |
|
Systolic blood pressure; mean (SD) |
126.5 (19.0) |
126.5 (19.3) |
126.9 (17.4) |
|
Diastolic blood pressure; mean (SD) |
72.0 (11.3) |
73.3 (10.5) |
65.0 (13.0) |
|
O2 saturation; mean (SD) |
95.4 (2.8) |
95.6 (2.2) |
94.7 (4.6) |
|
O2 saturation SD; mean (SD) |
2.6 (1.6) |
2.3 (1.0) |
4.1 (2.9) |
|
Initial Hypoxia; n (%) |
50 (11.4%) |
39 (10.7%) |
11 (15.5%) |
|
Respiratory rate; mean (SD) |
20.2 (4.5) |
20.0 (4.4) |
21.1 (4.7) |
|
Temperature; mean (SD) |
98.9 (1.2) |
98.8 (1.1) |
99.0 (1.2) |
|
Laboratory Findings* |
|||
|
WBC; mean (SD) [k/ul] |
7.4 (4.5) |
7.1 (4.1) |
9.0 (6.0) |
|
Hemoglobin; mean (SD) [g/dl] |
12.1 (2.1) |
12.2 (2.0) |
11.5 (2.6) |
|
Platelet; mean (SD) [k/ul] |
242.1 (103.4) |
253.7 (102.4) |
182.5 (87.4) |
|
Creatinine; mean (SD) [mg/dl] |
1.5 (2.0) |
1.4 (1.7) |
2.2 (2.8) |
|
Sodium mean (SD) [mmol/L] |
139.1 (4.4) |
138.9 (3.7) |
140.3 (7.0) |
|
Chloride mean (SD) [mmol/L] |
102.0 (5.1) |
101.9 (4.5) |
102.7 (7.5) |
|
Potassium mean (SD) [mmol/L] |
4.1 (0.6) |
4.1 (0.5) |
4.1 (0.7) |
|
eGFR; mean (SD) [ml/min] |
50.4 (16.0) |
51.9 (14.8) |
42.9 (19.3) |
|
Troponin T; mean (SD) [ng/mL] |
0.0 (0.1) |
0.0 (0.1) |
0.1 (0.1) |
|
High sensitivity troponin mean [ng/L] |
40.9 (114.0) |
36.9 (119.1) |
61.4 (80.5) |
|
BNP mean (SD) [pg/ml] |
2434.1 (8662.7) |
1881.2 (7863.9) |
5284.6 (11623.5) |
|
INR; mean (SD) |
1.0 (0.4) |
1.0 (0.3) |
1.1 (0.5) |
|
D-dimer; mean (SD) [mg/L FEU] |
3.1 (5.3) |
2.6 (4.0) |
5.7 (9.2) |
|
Ferritin; mean (SD) [ng/ml] |
994.6 (1236.2) |
915.1 (955.3) |
1404.1 (2134.5) |
|
Fibrinogen; mean (SD) [mg/dl] |
459.2 (123.9) |
461.9 (124.3) |
445.1 (121.5) |
|
Procalcitonin; mean (SD) [ng/ml] |
0.7 (4.5) |
0.4 (1.5) |
2.5 (10.5) |
|
Magnesium; mean (SD) [mg/dl] |
2.1 (0.3) |
2.1 (0.3) |
2.1 (0.3) |
|
Clinical Course |
|||
|
Hospital length of stay; mean (SD) |
16.2 (13.7) |
16.1 (14.1) |
17.0 (11.7) |
|
ICU Admission; n (%) |
167 (38.2%) |
122 (33.3%) |
45 (63.4%) |
|
Use of vasopressors; n (%) |
92 (21.1%) |
62 (16.9%) |
30 (42.3%) |
|
Hemodialysis; n (%) |
32 (7.3%) |
24 (6.6%) |
8 (11.3%) |
|
CPR; n (%) |
20 (4.6%) |
1 (0.3%) |
19 (26.8%) |
|
Discharge; n (%) |
|
||
|
Home |
265 (61.3%) |
260 (71.4%) |
5 (7.4%) |
|
Nursing Facility |
105 (24.3%) |
100 (27.5%) |
5 (7.4%) |
|
Expired in hospital |
55 (12.7%) |
0 (0.0%) |
55 (80.9%) |
|
Rehabilitation |
7 (1.6%) |
4 (1.1%) |
3 (4.4%) |
|
Other/Missing |
3 (0.007%) |
1 (0.002%) |
2 (0.03%) |
|
*Mean values for continuous variables indicate aggregate mean values for multiple values for the encounter. |
|||
Table 1: Clinical profile of patients hospitalized for COVID-19 by mortality status based on EHR data.
Table 1 describes the demographic profile of the full cohort with 366 patients who survived, and 71 patients who died. Patients who died were older and more often males with mean hospitalization for 17 days. They also had more comorbidities, and higher BNP, troponins, creatinine, ferritin, procalcitonin, d-dimer, and low platelets as compared to patients who survived.
Measurement of novel markers in hospitalized COVID-19 patients
Novel serum markers were measured from hospitalized patients with COVID-19. Patients who died had lower renalase values on average, and a trend for higher IL-1, KIM-1, and IFNs compared to patients who survived (Table 2).
|
Novel serum markers |
Total (n=437) |
Survived (n=366) |
Died |
|
(n=71) |
|||
|
Renalase mean (SD) [ng/ml] |
14046.5 (7960.5) |
14666.2 (8136.0) |
10852.0 (6098.2) |
|
Kidney injury molecule -1 (KIM-1) mean (SD) [ng/ml] |
122.8 (258.3) |
108.4 (221.0) |
197.1 (392.4) |
|
Interferon gamma (IFN-g) mean (SD) [pg/ml] |
302.3 (949.1) |
237.9 (617.0) |
634.4 (1868.6) |
|
Interferon alpha (IFN-a) mean (SD) [pg/ml] |
47.0 (147.4) |
41.3 (144.4) |
76.5 (159.6) |
|
Interferon lambda (IFN-l) mean (SD) [pg/ml] |
47.0 (147.4) |
41.3 (144.4) |
76.5 (159.6) |
|
Interleukin 1 (IL-1) mean (SD) [pg/ml] |
0.2 (0.9) |
0.2 (0.4) |
0.6 (2.0) |
|
Interleukin 6 (IL-6) mean (SD) [pg/ml] |
1614.7 (7312.1) |
1730.5 (7857.9) |
1017.9 (3259.5) |
|
Tumor necrosis factor alpha (TNF-a) mean (SD) [pg/ml] |
1614.7 (7312.1) |
1730.5 (7857.9) |
1017.9 (3259.5) |
Table 2: Profile of patients hospitalized with COVID-19 by mortality status and novel serum markers.
Predictors of mortality for hospitalized COVID-19
Combining traditional EHR abstracted data with novel serum markers, we used traditional and ML methods to identify predictors of mortality in the hospitalized patients. The results, shown in Table 3, indicate better performance when using the XGBoost models over traditional models.
|
Analytical Method |
Train AUC |
Validation AUC |
Test AUC |
|
A Priori Logistic Regression |
0.81 (0.75,0.87) |
0.81 (0.69,0.92) |
0.72 (0.62,0.82) |
|
Backwards Step Logistic Regression |
1 (1,1) |
1 (1,1) |
0.70 (0.55,0.77) |
|
XGBoost |
1 (0.98, 1) |
1 (0.98, 1) |
0.85 (0.77,0.93) |
Table 3: Comparison of AUC results using traditional and machine learning models to predict mortality in hospitalized patients with COVID-19. Confidence intervals for AUCs are in the parentheses.
Using the traditional logistic regression model with variables selected a-priori based on clinical observations (Supplemental Table S1), we identified age, patient sex and mean renalase to be significant predictors of mortality.
A backward step logistic regression identified clinical parameters (oxygen saturation) and several traditional laboratory parameters (hemoglobin, chloride, glomerular filtration rate, blood urea nitrogen, platelet count, BNP, troponins) in addition to renalase as predictors of mortality (Supplemental Table S2).
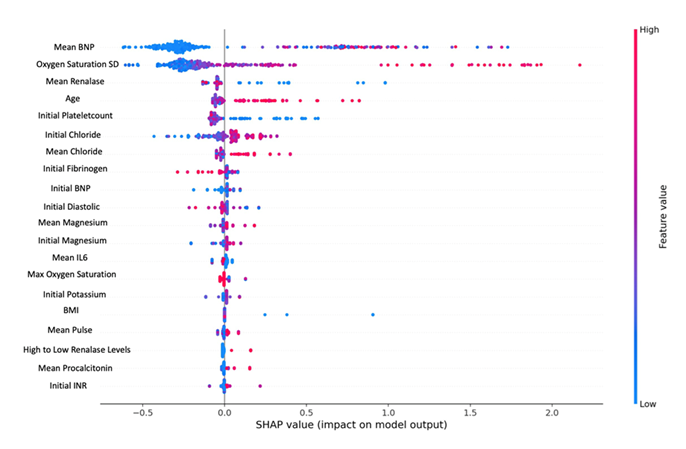
XGBoost model had the strongest performance based on AUC 0.85 (0.77,0.93) when tested. It identified similar variables as above with their relative importance as listed in (Figure 1), and showed high BNP being the most important predictor of mortality, followed by large standard deviation of oxygen saturation, renalase and advanced age.
Additional comparisons based on AUC-PR and calibration plots also indicate that the XG-Boost model has the best performance (see Supplemental Figure S2). Bootstrapping methods were used to generate different sample sets to test the models and generate confidence intervals. Summary data (Supplemental Table S4) shows lower for AUC for all models with XGBoost still performing the best.
Figure 1: SHAP value graph using XGBoost model identifies predictors of mortality in decreasing importance. Color indicates value (red=high or blue=low) and x-axis indicates survival (0) and death (1). For example, the blue dots on the left for BNP values indicate lower values are associated with survival (0), and on the left for renalase indicate association with death (1).
Prognostic value of Renalase as a predictor of COVID-19 mortality:
While the set of features are not identical in all the models due to the different ways they are trained, there is a large overlap in biomarkers and demographic features providing useful information about predicting mortality in hospitalized COVID-19 patients. Oxygen standard deviation, advanced age, elevated BNP, and low renalase appear to be significant predictors in at least two models. Low renalase emerged as a consistent predictor of mortality across all the tested models.
Profile of COVID-19 patients with serial samples available
We next assessed the dynamic changes in renalase in relation to outcomes in COVID-19 patients. At baseline renalase levels vary from 4 to 100 mg/L among in healthy non-hospitalized individuals. Based on in-vivo data, we hypothesized that changes in renalase level in response to COVID-19 from baseline within the same individual would predict outcomes. To assess this hypothesis, we tested serial renalase in our study sub-cohort (n=124) who had at least 3 serial samples available. Supplemental Table S3 notes the profile of these patients. Twenty-one patients died and were similar in age, sex, and race distribution as the overall cohort.
Serial course of renalase over hospital course in COVID-19
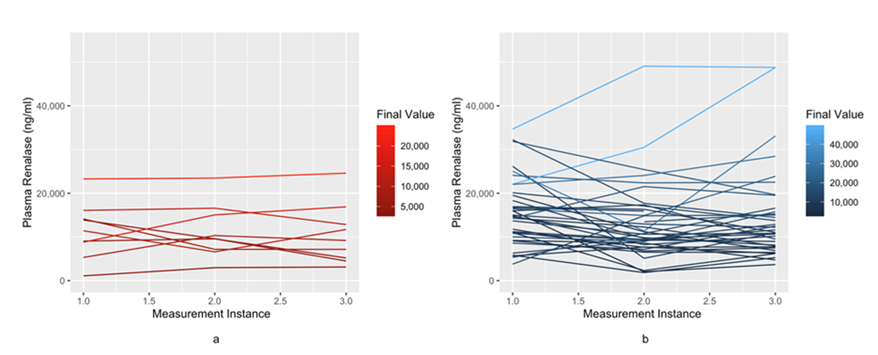
Figure 2 provides visualization of serial renalase values in patients who survived versus died. Patients who had low renalase levels and remained low tended to do poorly compared to those patients whose renalase values stayed high over their hospitalization (P-value of <0.001), indicating that different trajectories experienced different levels of mortality.
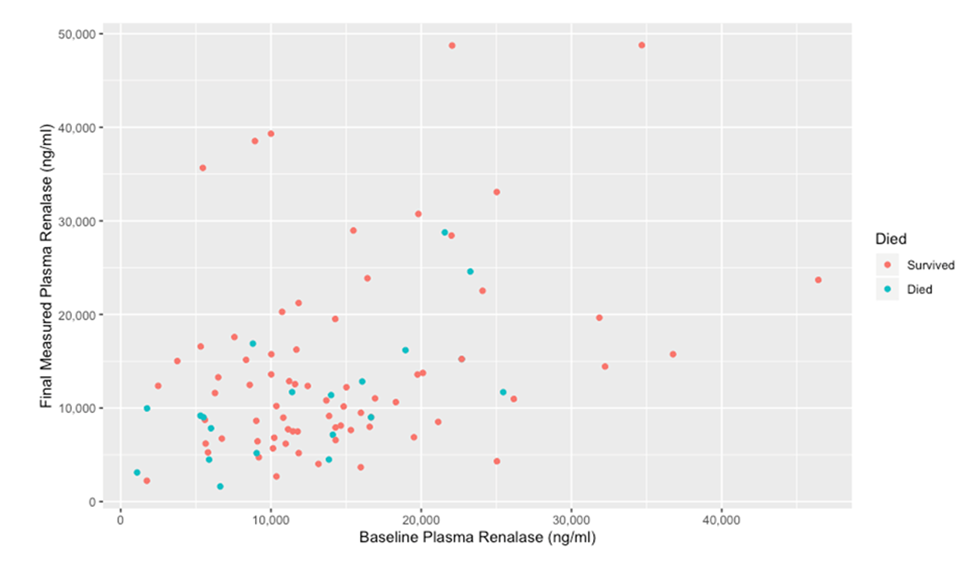
We then visualized comparisons between the baseline and final renalase for patients depending on survival status.
Figure 3 shows patients who died had lower baseline and final renalase values on average compared to those who survived.
Next, we visualized the relationship of renalase trajectory relative to that of other biomarkers consistently identified as predictors of mortality in our models:
a) Relationship between renalase and markers of cardiac injury
In the XGBoost model, a high BNP, a marker of cardiac strain, appeared to be the most important predictor of mortality. Supplemental Figure S1A compares them by quartiles. Patients with lowest (Q1) RNLS-highest (Q4) BNP quartile had significantly higher mortality than patients with high (Q4) RNLS- low (Q1) BNP quartile (P-value of 0.003) (Supplemental Figure S1A). This was true even when comparing the trajectory of these markers - patients with low renalase and high BNP at the end of their hospitalization had worse mortality compared to patients with high renalase and low BNP at the end of their hospitalization (P-value <0.0001).
Renalase showed a similar relationship with high-sensitivity troponin, a marker of cardiac injury. Although not significant in the XGBoost model, troponins were found to predict mortality in traditional models. Supplemental Figure S1B heatmap showed that in serial samples, patients with Q1RNLS-Q4Troponin had higher mortality than patients with high renalase and lower troponin (Q4RNLS-Q1troponin) quartile (P-value of 0.002).
b) Relationship between renalase and inflammatory markers
The relationship is also less clear when comparing renalase with inflammatory markers such as IL-1, IL-6 and IFN. Supplemental Figure S1C shows comparison of mortality in patients with low (Q1) RNLS- high (Q4) IL-6 to be significantly different than in patients with high (Q4) RNLS-low (Q1) IL-6 (p value=0.02). However, no difference was found for comparison of similar cohorts for IL-1 (p value=0.58) and IFN (p value=0.07) Supplemental Figure S1D and S1E.
c) Relationship between renalase and platelets
Finally, in the XGBoost model, platelet count also appeared to be an important predictor of mortality. Supplemental Figure S1F compares different pairs of trajectories for renalase and platelet count values indicate patients with lower (Q1) renalase values and lower platelet counts (Q1) had the highest rates of death compared to those with high (Q4) renalase-high (Q4) platelets (p =0.005).
Discussion
In this single site study evaluating predictors of mortality in patients hospitalized with COVID-19, renalase was identified as a novel independent predictor of mortality - consistently implicated using both traditional statistical methods based on a priori knowledge, as well as when applying more agnostic ML methods. The XGBoost model was identified as the best performing model with AUC of 0.87 in predicting mortality from COVID-19. Using this model, elevated BNP, greater change in oxygen saturation, low renalase, and advanced age were found to be the strongest predictors of mortality in order among hospitalized patients. A significant relationship between the trajectory of renalase and markers of thromboembolism and cardiac strain sheds light on the pathophysiology of renalase in host response to COVID-19.
Advanced age and hypoxia are known hallmark predictors of COVID-19 severity [17, 18]. Similarly, cardiac markers such as elevated BNP and troponin have been associated with adverse outcomes in severe COVID-19 [19,20]. The reasons appear multifactorial - SARS-COV2 virus invades cardiac myocytes via the angiotensin-converting enzyme 2 (ACE-2) receptor, abundantly present through heart and blood vessels [21]. The injury degrades both the cardiac myocytes as well as the ACE-2 receptor in the coronary microvasculature. The resultant overactivation of the renin-angiotensin-aldosterone (RAAS) pathway causes widespread coronary microvascular dysfunction, inflammation-induced endothelial apoptosis, vascular permeability, prothrombosis, and an excess catecholamine state implicated in severe COVID-19 [22-24]. There is also evidence that systemic microvascular dysfunction and tissue hypoxemia causes acute pulmonary hypertension, right ventricular stress as evident by echocardiogram and on autopsies findings leading to BNP release [25,26]. The XGBoost model supports this theory with higher mortality prediction linked with mean BNP values, as a marker of cardiac strain as compared to troponin, a marker of myocardial injury. A novel contribution from our study is identifying low plasma renalase level as the most consistent independent predictor of mortality in all our tested models, and the second most important laboratory predictor using the XGBoost method. Renalase was originally discovered as a protein to explain the high cardiovascular burden observed in persons with chronic kidney disease [27]. We noted low renalase to be a predictor of mortality in COVID-19 as well, both when assessed at baseline as well as serially. Patients whose trajectory for renalase remained low had significantly higher mortality as opposed to patients who produced high renalase levels during their hospitalization. Similar trajectories were observed in relation to renalase and other peptides – most significantly in relation to markers of cardiac strain (BNP, troponin) and thromboembolism (platelets). Interestingly, renalase levels did not correlate with inflammatory cytokines such as IL-1 and IL-6, highlighting additional protective pathways. A high BNP and low renalase were linked with worse prognosis than the inverse. In non-COVID heart failure patients, renalase has been shown to add discriminatory and prognostic value for ischemia protection to BNP, both thought to be released in response to an overstimulated catecholamine state as well as RAAS activation [28-30]. In addition, low-tissue oxygenation state in heart failure, which is also seen in COVID-19, triggers the release of hypoxia inducible factor (HIF 1-alpha), a known activator of renalase secretion and transcription. Renalase initiates protective receptor signal transduction mechanisms (STAT3), mitogen activated protein kinase (MAPK) and protein kinase B, [31] and inhibits profibrotic gene expression playing an important role in cardiac remodeling [32, 33]. Addition of recombinant renalase in mice models of heart failure has been shown to decrease myocardial necrosis and improve ejection fraction [34]. Conversely, low renalase production has been shown to worsen heart failure, indicated by rising BNP, and confirmed by increased myocardial apoptosis [34]. We also observed an association between renalase, platelets and mortality. Thrombo-inflammation and micro-embolization seen in severe COVID-19 has been linked with endothelial injury resulting in widespread platelet consumption [35]. Renalase appears to impart endothelial protection by activating the PMCA4B, stabilizing cell membrane, metabolizing catecholamines, and reducing cytokine production [36]. Renalase also appears to show cell protection systemically in animal models [34, 37, 38]. Second, the RAAS imbalance in COVID-19 causing angiotensin II activation of AT1R also triggers apoptosis nuclear factor (NF-kb) that induces release of inflammatory cytokines (IL-1, IL-6) and thrombotic factors (platelet derived growth factor) [39]. This pathway triggers compensatory activation of renalase peptide, potentially explaining the link between renalase and platelets [40].
Our study highlights the role of ML methods, particularly XGBoost, by showing strong predictive results. With a high AUC of 0.85, the XGBoost model effectively differentiated patients who would experience a mortality event from those who would not, augmenting the knowledge we have gained with traditional models.
Our results should be interpreted considering certain limitations. This was a single site retrospective study in the first wave of COVID-19; hence the generalizability of our findings should be tested in larger varied cohorts and with COVID-19 infections with newer strains. Testing of novel serum markers in a national cohort however is logistically more difficult and our data adds strength to traditional electronic data. Second, serial samples were available only in a small number of patients. Serial sampling itself may indicate more severe illness, and therefore may not represent the overall population. However, we believe a minimal effect of this bias as excluded patients were similar in profile. Third, since our data was pre-vaccination era it is unclear how immunization would influence our results.
Conclusion
Machine learning methods augment traditional statistical methods in identifying novel predictors of mortality with COVID-19. Renalase was identified as a consistent and independent predictor of mortality in patients hospitalized with COVID-19. The trajectory of renalase, especially in conjunction with other markers of cardiac and endothelial injury, should be explored in prospective studies.
Competing Interests
G Desir is a named inventor on several issued patents related to the discovery and therapeutic use of renalase. Renalase is licensed to Bessor Pharma, and G Desir holds an equity position in Bessor and its subsidiary Personal Therapeutics. J El-Khoury has received grants from Siemens Healthineers, Bioporto, and IDEXX and honoraria from the Association of Diagnostic and Laboratory Medicine. He acts as a consultant for Siemens Healthineers and holds board positions in Clinical Chemistry and Association for Diagnostic and Laboratory medicine. He has also received free equipment from Bruker. B Safdar is recipient of National Institute of Health funding for role of renalase in COVID-19 (1OTHL56812-01). The interests had no influence on study design, data collection, analysis, interpretation of study results, the writing of the report or the decision to submit the report for publication. There are no other competing interests to declare.
Acknowledgements
We want to thank the selflessly included patients and analytical unit platform staff for their help with this study. We would also like to thank Dana Lee for the helping with formatting and submission of this manuscript.
Preprint Disclosure
A preprint version of this manuscript is available on Research Square (DOI: https://doi.org/10.21203/rs.3.rs-2492699/v1, https://www.researchsquare.com/article/rs-2492699/v1)
Author Contribution Statements
B.S., M.S. D.B, G.D. and G.D. designed the study; B.S., A.N. G.D. J.K. M.S. M.W. and G.D. collected the data; X.G. carried out experiments; M.S., D.B, Y.D. and J.D. analyzed the data; M.S. and A.T. made the figures; B.S., M.S., D.B, A.N., Y.D., G.D., J.D., X.G., A.T., M.W., and G.D. drafted and revised the paper; All authors have critically revised the manuscript, takes responsibility for the integrity and accuracy analyses of the data, and have approved the manuscript's final version of the manuscript.
Supplementary File Link:
https://www.fortunejournals.com/supply/JBB_10071.pdf
References
- Organization WH. https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19 (2023).
- Samprathi M, Jayashree M. Biomarkers in COVID-19: an up-to-date review. Front Pediatr 8 (2020): 607647.
- Sinha P, Matthay MA, Calfee CS. Is a “cytokine storm" relevant to COVID-19? JAMA Intern Med 9 (2020): 1152-1154.
- Zhang NH, Cheng YC, Luo R, et al. Recovery of new-onset kidney disease in COVID-19 patients discharged from hospital. BMC Infect Dis 1 (2021): 397.
- Safdar B, Wang M, Guo X, et al. Association of renalase with clinical outcomes in hospitalized patients with COVID-19. PLoS One 3 (2022): e0264178.
- Serwin N, Cecerska-Heryc E, Pius-Sadowska E, et al. Renal and inflammation markers-renalase, cystatin C, and NGAL levels in asymptomatic and symptomatic SARS-CoV-2 infection in a one-month follow-up study. Diagnostics (Basel) 12 (2022): 1.
- Guo X, Chen R, Chen T, et al. Renalase ameliorates AKI by altering mitochondrial function to induce cellular repair mechanisms. American Society of Nephrology (2023).
- Doll S, Proneth B, Tyurina YY, et al. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat Chem Biol 1 (2017): 91-98.
- Arora T SM, Alausa J, Subair L, et al. The Yale Department of Medicine COVID-19 Data Explorer and Repository (DOM-CovX): an innovative approach to promoting collaborative scholarship during a pandemic. (2021).
- Chang J, Guo X, Rao V, et al. Identification of two forms of human plasma renalase, and their association with all-cause mortality. Kidney Int Rep 3 (2020): 362-368.
- Dong Y, Peng CY. Principled missing data methods for researchers. Springerplus 1 (2013): 222.
- Bertsimas D PC, Zhuo YD. From predictive methods to missing data imputation: an optimization approach. The Journal of Machien Learning Research 1 (2017): 7133-7171.
- TCaC. Xgboost: A scalable tree boosting system. In: Mining IPotnASICoKDaD, editor. New York: Association for Computing Machinery (2016).
- Olson RS, Cava W, Mustahsan Z, et al. Data-driven advice for applying machine learning to bioinformatics problems. Pac Symp Biocomput 23 (2018): 192-203.
- Lundberg SM, Lee SI. A unified approach to interpreting model predictions. Advances in neural information processing systems 30 (2017).
- JA MPaN. Generalized Linear Models. In: Hall C, editor. Monographs on Statistics and Applied Probability Series (1989).
- Ho FK, Petermann-Rocha F, Gray SR, et al. Is older age associated with COVID-19 mortality in the absence of other risk factors? General population cohort study of 470,034 participants. PLoS One 11 (2020): e0241824.
- Xie J, Covassin N, Fan Z, et al. Association between hypoxemia and mortality in patients with COVID-19. Mayo Clin Proc 6 (2020): 1138-1147.
- Qin JJ, Cheng X, Zhou F, et al. Redefining cardiac biomarkers in predicting mortality of inpatients with COVID-19. Hypertension 4 (2020): 1104-1112.
- Guo T, Fan Y, Chen M, et al. Cardiovascular implications of fatal outcomes of patients with Coronavirus Disease 2019 (COVID-19). JAMA Cardiol 7 (2020): 811-818.
- Yin J, Wang S, Liu Y. Coronary microvascular dysfunction pathophysiology in COVID-19. Microcirculation 7 (2021): e12718.
- Oudit GY, Kassiri Z, Jiang C, et al. SARS-coronavirus modulation of myocardial ACE2 expression and inflammation in patients with SARS. Eur J Clin Invest 7 (2009): 618-625.
- Chirinos JA, Cohen JB, Zhao L, et al. Clinical and proteomic correlates of plasma ACE2 (angiotensin-converting enzyme 2) in human heart failure. Hypertension 5 (2020): 1526-1536.
- Ma M, Xu Y, Su Y, et al. Single-cell transcriptome analysis decipher new potential regulation mechanism of ACE2 and NPs signaling among heart failure patients infected with SARS-CoV-2. Front Cardiovasc Med 8 (2021): 628885.
- Caliskan M, Baycan OF, Celik FB, et al. Coronary microvascular dysfunction is common in patients hospitalized with COVID-19 infection. Microcirculation 5 (2022): e12757.
- Deng Q, Hu B, Zhang Y, et al. Suspected myocardial injury in patients with COVID-19: Evidence from front-line clinical observation in Wuhan, China. Int J Cardiol 311 (2020): 116-121.
- Xu J, Li G, Wang P, et al. Renalase is a novel, soluble monoamine oxidase that regulates cardiac function and blood pressure. J Clin Invest 5 (2005): 1275-1280.
- Stojanovic D, Mitic V, Stojanovic M, et al. The discriminatory ability of renalase and biomarkers of cardiac remodeling for the prediction of ischemia in chronic heart failure patients with the regard to the ejection fraction. Front Cardiovasc Med 8 (2021): 691513.
- Han P, Sun H, Xu Y, et al. Lisinopril protects against the adriamycin nephropathy and reverses the renalase reduction: potential role of renalase in adriamycin nephropathy. Kidney Blood Press 5 (2013): 295-304.
- Richards AM. The renin-angiotensin-aldosterone system and the cardiac natriuretic peptides. Heart 3 (1996): 36-44.
- Wang Y, Safirstein R, Velazquez H, et al. Extracellular renalase protects cells and organs by outside-in signalling. J Cell Mol Med 7 (2017): 1260-1265.
- Stojanovic D, Mitic V, Stojanovic M, et al. The partnership between renalase and ejection fraction as a risk factor for increased cardiac remodeling biomarkers in chronic heart failure patients. Curr Med Res Opin 6 (2020): 909-919.
- Du M, Huang K, Huang D, et al. Renalase is a novel target gene of hypoxia-inducible factor-1 in protection against cardiac ischaemia-reperfusion injury. Cardiovasc Res 2 (2015): 182-191.
- Li X, Xie Z, Lin M, et al. Renalase protects the cardiomyocytes of Sprague-Dawley rats against ischemia and reperfusion injury by reducing myocardial cell necrosis and apoptosis. Kidney Blood Press 3 (2015): 215-222.
- Taus F, Salvagno G, Cane S, et al. Platelets promote thromboinflammation in SARS-CoV-2 pneumonia. Arterioscler Thromb Vasc Biol 12 (2020): 2975-2989.
- Kolodecik TR, Reed AM, Date K, et al. The serum protein renalase reduces injury in experimental pancreatitis. J Biol Chem 51 (2017): 21047-21059.
- Lee HT, Kim JY, Kim M, et al. Renalase protects against ischemic AKI. J Am Soc Nephrol 3 (2013): 445-455.
- Zhang T, Gu J, Guo J, et al. Renalase attenuates mouse fatty liver ischemia/reperfusion injury through mitigating oxidative stress and mitochondrial damage via activating SIRT1. Oxid Med Cell Longev 2019 (2019): 7534285.
- Okamoto H, Ichikawa N. The pivotal role of the angiotensin-II-NF-kappaB axis in the development of COVID-19 pathophysiology. Hypertens Res 1 (2021): 126-128.
- Aoki K, Yanazawa K, Tokinoya K, et al. Renalase is localized to the small intestine crypt and expressed upon the activation of NF-kappaB p65 in mice model of fasting-induced oxidative stress. Life Sci 267 (2021): 118904.


 Impact Factor: * 5.3
Impact Factor: * 5.3 Acceptance Rate: 75.63%
Acceptance Rate: 75.63%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 