The Assessment of Methods for Preimplantation Genetic Testing for Aneuploidies Using a Universal Parameter: Implications for Costs and Mosaicism Detection
Alexander Belyaev1*, Maria Tofilo2, Sergey Popov2, Ilya Mazunin2, Dmitry Fomin3
1AB Vector LLC, San Diego, CA, USA
2Medical Genomics Laboratory, Tver, Russia
3Network of Medical Centers «Fomin Clinics», Moscow, Russia
*Corresponding author: Alexander Belyaev, AB Vector LLC, 3830 Valley Center Dr., San Diego 92130, CA, USA.
Received: November 16, 2023; Accepted: January 5, 2024; Published: January 25, 2024
Article Information
Citation: Alexander Belyaev, Maria Tofilo, Sergey Popov, Ilya Mazunin, Dmitry Fomin. The Assessment of Methods for Preimplantation Genetic Testing for Aneuploidies Using a Universal Parameter: Implications for Costs and Mosaicism Detection. Journal of Biotechnology and Biomedicine. 7 (2024): 47-59.
DOI: 10.26502/jbb.2642-91280126
View / Download Pdf Share at FacebookAbstract
Preimplantation genetic testing for aneuploidies (PGT-A) is used to increase live birth rates following in vitro fertilization. The assessment of different PGT-A methods to date has relied on non-universal parameters, e.g., sensitivity and specificity, that are individually stipulated for each study and typically performed using arbitrarily selected cell lines. Here we present an alternative approach that is based on an assessment of the median noise in a large dataset of routine clinical samples. Raw sequencing data obtained during PGT-A testing of 973 trophectoderm biopsies was used for comparison of two methods: VeriSeq PGS (Illumina) and AB-PGT (AB Vector). Three times less median noise was a feature of the AB-PGT method, thereby allowing the number of multiplexed samples per sequencing run to be increased from 24 with VeriSeq PGS to 72 with AB-PGT, thus effectively reducing the price per sample without compromising data quality. The improvement is attributed to a novel SuperDOP whole genome amplification technology combined with a simplified PGT-A protocol. We show that the median noise level associated with a large dataset of biopsies is a simple, universal metric for the assessment of PGT-A methods, which has implications for other screening methods, the detection of mosaicisms, and the improvement of fertility clinics’ practices.
Article Details
Introduction
Aneuploidy is the primary reason embryos in both natural and assisted reproductive cycles fail to result in a healthy pregnancy [1, 2]. Analysis of the ploidy state of the genome is often referred to as copy number variation (CNV) analysis [3–5]. Illumina’s array comparative genomic hybridization (aCGH) platform was the first widely used 24-chromosome PGT-A method to determine the ploidy state of an embryo [6]. Later, a more powerful next generation sequencing (NGS) platform gained popularity and became the industry standard [7, 8]. With this method, samples are prepared using Illumina’s SurePlex and VeriSeq PGS kits, sequenced on the Illumina’s MiSeq benchtop sequencer, and CNV analysis is performed using BlueFuse Multi, Illumina software. Recently, the Embryomap kit became available from the same manufacturer, though peer review data on its performance has yet to be reported. All methods require whole genome amplification (WGA) to generate sufficient DNA for analysis, with PCR-based methods providing better uniformity of genome coverage for PGT-A than other methods [9–11]. In PCR-based methods that do not involve adapter ligation, primers with non-degenerate 5’-portion and degenerate nucleotides in the 3’-portion are used. First, several low-stringency (LS) PCR cycles are performed in which the primer anneals to a template at multiple sites via the degenerate 3’-portion, aka “random” priming. Then the temperature is increased to allow primer extension, and, after several such cycles, amplicons are subjected to high-stringency (HS) PCR cycles at a much higher annealing temperature [12]. This allows exponential amplification using the non-degenerate portions of primers and excludes “random” priming.
With the development of a new method, there is a need for comparison to an accepted gold standard [7]. For example, the more recently developed Ion Single Seq kit (Thermo Fisher Scientific, USA) was compared with the industry standard VeriSeq PGS kit and shown to perform similarly [4, 5]. In our earlier study, the VeriSeq PGS kit (Illumina) was compared with the AB-PGT kit (AB Vector) using aneuploid cell lines and discarded blastocysts [13]. High concordance between the kits in the detection of aneuploidies was observed. Such concordance studies can establish that kits perform similarly, but their efficiency in detecting aberrations is not compared. To this end, sensitivity and specificity studies are performed to compare different methods [4, 5]. However, these comparisons were done on cell lines and involved a subjective element–manual identification of aberrations. Indeed, in screening tests such as PGT-A, sensitivity and specificity parameters are not easily defined and often involve subjective assessments that can be ambiguous [14, 15]. Ideally, a clinical study should serve as a universal approach for an assessment of PGT-A methods. However, clinical studies are even more complex [16] and “prone to pitfalls that preclude the obtention of definitive conclusions to guide an evidence-based approach for such a challenging population” [17]. As a result, despite numerous clinical studies, key PGT-A aspects remain controversial [18–21].
In this study, we pioneered a hybrid approach that makes use of pre-existing sequencing data from 973 biopsies obtained during routine PGT-A testing in a clinical setting. Since the sequencing data for the PGT-A methods used are in the same format, they can be analyzed using the same software proprietary to AB Vector. The large number of routinely processed PGT-A samples allowed for definitive conclusions regarding the performance of both methods using the median noise parameter. We show that although there is a large spread in the quality of individual biopsies, the median noise level across the dataset is a simple, universal metric that can be used to assess the performance of different PGT-A methods. As “CNVs are expected to have a tremendous impact on screening, diagnosis, prognosis, and monitoring of several disorders, including cancer and cardiovascular disease" [22], we suggest that our approach may apply not only to PGT-A but to other CNV-based tests as well.
Materials and Methods
Study design
This study was approved by the Ethics Review Committee of the Network of Medical Centers «Fomin Clinics», Moscow, Russian Federation. The study was performed in four phases. In the first phase, we analyzed the noise levels of 973 biopsies that were processed using VeriSeq PGS or AB-PGT kits and determined the median noise level characteristic for each kit contingent upon the number of MiSeq reads per sample. In the second phase, we simulated different levels of noise in an aneuploid sample and studied the noise impact on the detection of aneuploidies. In the third phase, we simulated mosaicisms by mixing reads from euploid and aneuploid samples and investigated how noise levels affect the detection of mosaicisms of varying amplitude in terms of copy number. In the fourth phase, we compared the performance of CNVector (AB Vector) and BlueFuse Multi (Illumina) software on the mosaic samples.
Patients
The study is representative of typical healthy fertility clinic patients. The median age of patients was 37.6 ± 5.7 standard deviation (SD) in the group tested with the AB Vector PGT-A kit and 38.0 ± 6.2 SD in the group tested with the Illumina Veriseq PGS kit. For both groups, patients underwent routine oocyte retrieval, followed by intracytoplasmic sperm injection of MII oocytes. The injected oocytes were cultured to the blastocyst stage, and trophectoderm biopsies were taken and frozen. After the biopsy, blastocysts were vitrified and stored in liquid nitrogen until the PGT-A data became available and implantation decisions regarding specific embryos were made.
Sample preparation and sequencing
Biopsies were processed with AB Vector or Illumina kits according to the manufacturer’s instructions. In brief, for the Illumina workflow, the biopsies were first processed using the SurePlex kit. This involved cell lysis and DNA extraction, pre-amplification comprising 12 LS PCR cycles, and a subsequent separate amplification reaction comprising 14 HS PCR cycles. The success of the amplification was controlled using agarose gel electrophoresis. Next, SurePlex samples were processed using the VeriSeq PGS kit. In brief, amplified SurePlex DNA products were quantitated using the Qubit method and fragmented in a tagmentation reaction using a hyperactive variant of the TN5 transposase. The fragmented DNA was double-tagged by a 19-nucleotide DNA sequence that was attached to the ends of the fragments by the transposase, thus facilitating further rounds of amplification. Using these tags, p5 and p7 indexes were added to these fragments in PCR (indexing PCR). The PCR products were purified from indexing primers on AMPure XP magnetic beads (Beckman-Coulter) and subjected to a normalization step on proprietary magnetic beads (Illumina). Finally, the libraries were pooled at equimolar concentrations for sequencing on MiSeq. As recommended, dual-indexed sequencing on a paired-end flow cell with 36 cycles Read 1 was implemented, and Read 2 was omitted.
The AB Vector sample preparation workflow does not require two kits; a single AB-PGT kit was used according to the manufacturer’s instructions. Though the start and end points of Illumina and AB Vector workflows are very similar, the latter requires fewer steps. The agarose gel electrophoresis, tagmentation, and normalization steps were omitted. The AB Vector workflow comprises cell lysis and DNA extraction, proprietary SuperDOP WGA, purification from primers and large DNA fragments on the AMPure XP magnetic beads (sizing), indexing PCR, further sizing, and finally pooling libraries with the size of DNA fragments of about 400–550 b.p. for sequencing on MiSeq. The MiSeq setup was the same as with VeriSeq PGS, except that Read 1 was 72 cycles.
CNV analysis
For the comparison of two dissimilar sample preparation methods, it is pivotal to have CNV analysis software that works equally well with both. To this end, CNVector software was developed that comprises exactly the same algorithms for both methods, except reference data used for removing bias inherent in the PGT-A method differed for AB Vector and Illumina samples since the biases generated by each method differed.
FASTQ files generated after sequencing on MiSeq of AB-PGT or VeriSeq PGS samples were processed using CNVector software. Reads were demultiplexed into individual samples according to double indexing and aligned to the human genome hg19. We removed unmapped reads, duplicate reads, and reads with low mapping scores. The genome was divided into 2,000 bins, each containing an equal number of reads. Variable bin width is required for a constant number of reads per bin since the density of reads varies across the genome due to bias. The median bin width was 1.6 Mb, with a minimum of three bins, or ∼ 4.8 Mb median requirement for detecting an aberration. Filtered reads from each sample were then mapped into the corresponding chromosome interval or bin. The count data in each bin was normalized using the reference data to remove the bias created in either the AB Vector or Illumina sample preparation methods. The normalized bin counts were then re-expressed as copy number (CN) by assuming that the median autosomal read count, after removing autosomes with aneuploidies and mosaicisms, corresponds to a CN of two.
After binning and normalization, CNVector software performs a single median filtering operation, followed by a sweep of the genome to identify statistically significant jumps in the CN across neighboring bins. The genome is then broken into regions at the jump locations (if there are N jumps, there are N+1 regions). The maximum size of a region is a single chromosome, so there is always a region boundary between chromosomes. However, region boundaries can also occur within a given chromosome in the case of a discontinuous jump in CN within a chromosome due to an abnormality. The CN is assumed to be constant within a region but can differ between adjacent regions. A digital 1-2-1 filter is applied successively ten times to the raw binned data within each individual region (i.e., without filtering across region boundaries). This workflow suppresses random noise that is due to statistical fluctuations while ensuring there is no smearing of the CN across region boundaries.
Embryos are diagnosed as abnormal or aneuploid if the mean CN in any region deviates from the normal CN by a statistically significant value. In the case of mosaicisms, for which the CN in a region is not an integer value, the average CN value of the region compared to the standard deviation of the noise over the genome plays a key role. The standard deviation in copy units is calculated by the CNVector program according to the formula:
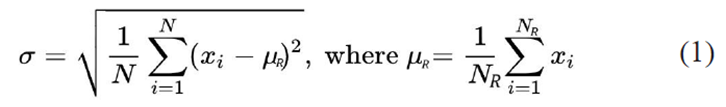
Here, N is the total number of bins spanning the genome (N = 2000 for CNVector by default), xi is the CN of the i-th bin, and NR is the number of bins in a region with a mean CN equal to mR. The standard deviation, sigma, provides a distribution-independent overall measure of the noise (s noise) in units of CN for a given sample after the CN data has been broken into regions and filtered.
The noise in a given sample consists of different sources, such as Poisson noise due to fluctuations in the number of reads per bin and systematic noise due to biopsy quality, the PGT-A method, and sequencing. Although the Poisson noise follows a Gaussian distribution in the limit of a large number of reads per bin, there is no guarantee or expectation that the systematic noise follows a Gaussian distribution. This means that the usual sigma confidence intervals for Gaussian distributions do not apply, and the underlying distribution for the systematic noise is assumed to be more heavy-tailed than Gaussian. For this reason, a conservative threshold is adopted for CNV detection. Thresholds (N sigma) for probable “observation of” (2.8 sigma) and practically certain “evidence for” (5 sigma), equivalent to the term “discovery of,” are standard practice in situations where the sources of systematic noise are complex and uncharacterized [23]. Here, N sigma thresholds are used to flag chromosomal abnormalities. N sigma determines the number of standard deviations (sigmas) that the mR of a given region deviates from normality. For instance, in the case of autosomes, the normal (euploid) CN value is two. Hence, within a given region, N sigma = |2-mR|/s. Here, s and mR are both in units of copy number, and the vertical brackets denote an absolute value, implying N sigma is always positive. The division by s in the formula for N sigma effectively generates a unit conversion from units of CN to sigmas.
By default, CNVector software flags a given region as probable detection of an abnormality if 3 ≤ N sigma ≤ 6, which is slightly more stringent than the “evidence of” threshold used in the literature. These regions are marked with yellow points in the graphical CNV output. Regions with N sigma > 6 indicate a practically certain detection of an abnormality. Such regions are marked with red points in the graphical CNV output. Normal regions of the genome are considered to have N sigma < 3 and are marked with blue points. In addition to graphical outputs for the entire genome and for individual chromosomes, quantitative information about mR and N sigma is presented in tabular form.
Results
In silico comparison of PGT-A methods
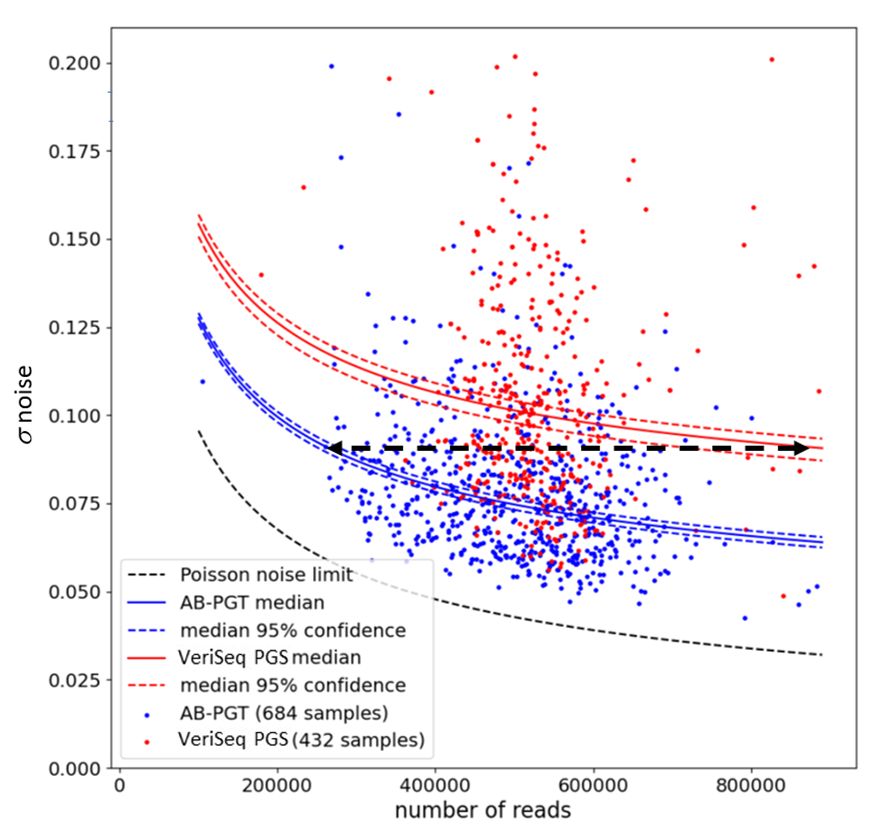
We show that s noise can be used as a metric for measuring the accuracy of different sample preparation PGT-A methods and for assessing the quality of individual biopsy samples. The former requires a large dataset of samples because of large differences in their quality. In this case, a median s noise value for a PGT-A method can be computed as a function of the number of reads per sample. Raw sequencing data from 584 AB-PGT samples and 389 VeriSeq PGS samples were processed using CNVector software (Fig. 1), and s noise was calculated for each sample using CNVector software that was adapted for each method. Poisson noise limit is a hypothetical scenario wherein a kit, a biopsy quality, and a sequencing method are ideal and generate no noise, thus all the noise is stipulated by the limited number of reads. The arrowed dash line indicates that the same median noise was observed with 260,000 reads using the AB-PGT kit as with 870,000 reads using the VeriSeq PGS kit.
Fig. 1: Median noise levels with AB-PGT and VeriSeq PGS kits contingent upon the number of reads. Libraries that were prepared using either method were sequenced on MiSeq. FASTQ files from both types of libraries were processed using CNVector software that was adapted for each method. Poisson noise limit is a hypothetical scenario wherein a kit, a biopsy quality, and a sequencing method are ideal and generate no noise, thus all the noise is stipulated by the limited number of reads. The arrowed dash line indicates that the same median noise was observed with 260,000 reads using the AB-PGT kit as with 870,000 reads using the VeriSeq PGS kit.
The noise for each AB-PGT sample (blue points) and Veriseq PGS sample (red points) was plotted against the number of filtered reads in the samples. The dashed black line shows the theoretical lower noise limit, which is due to statistical fluctuations in the number of reads per bin. Since these statistical fluctuations follow a Poisson distribution, the theoretical lower limit can be calculated. Although some samples do approach the theoretical limit, most points lie well above it for both the AB-PGT and Veriseq PGS methods.
The solid blue and red curves in Fig. 1 show the median s noise, described above, for the AB-PGT and Veriseq PGS methods, respectively. To estimate if the median noise levels differ significantly, confidence intervals were obtained via bootstrapping [24]. Bootstrapping assumes the data points accurately represent the underlying probability distribution and samples them with replacements, generating multiple synthetic data sets. For the 95% confidence interval (dashed blue and red curves), ± 0.0016 variation for AB-PGT and ± 0.0034 variation for Veriseq PGS medians in units of CN were observed. With Ds ∼ 0.03 between the medians in units of CN, there was a clear separation of the medians (P-value << 0.05), indicating significantly less noise with the AB-PGT method. Notably, the same median noise level was observed with 260,000 reads using the AB-PGT kit as with 870,000 reads using the Veriseq PGS kit (Fig. 1). This implies a threefold increase in the number of AB-PGT samples that can be multiplexed per sequencing run without increasing the median s noise value, thus equating to a maximum of 72 samples, as opposed to multiplexing a maximum of 24 VeriSeq PGS samples per run.
Mosaicisms detection is contingent upon the noise level
Starting from a low-noise sample, a higher noise level can be generated by downsampling, i.e., randomly reducing the number of reads in silico for the sample (Fig. 2).
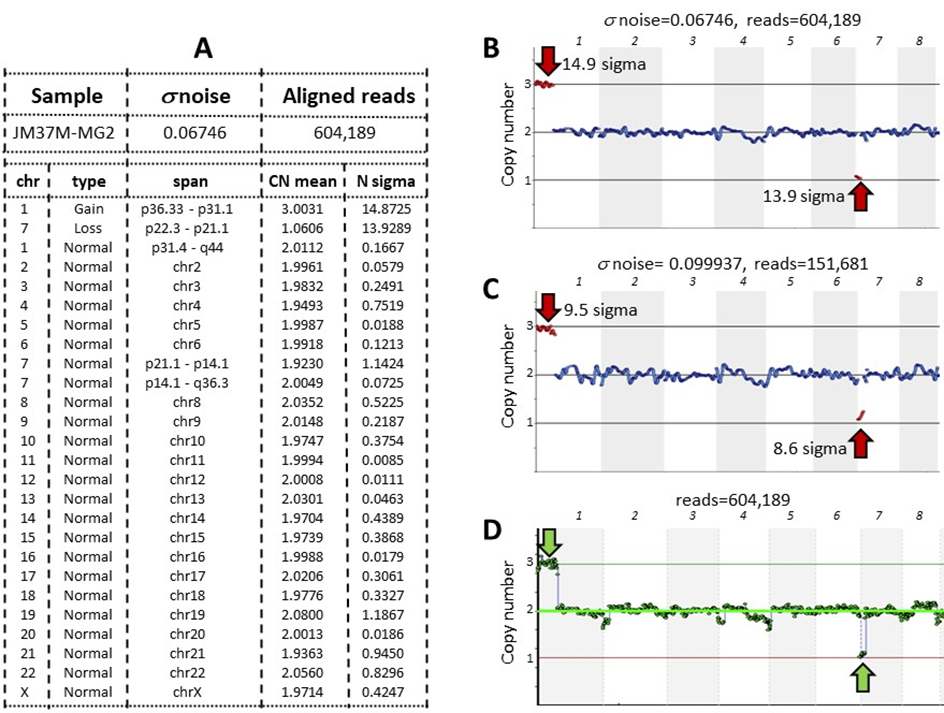
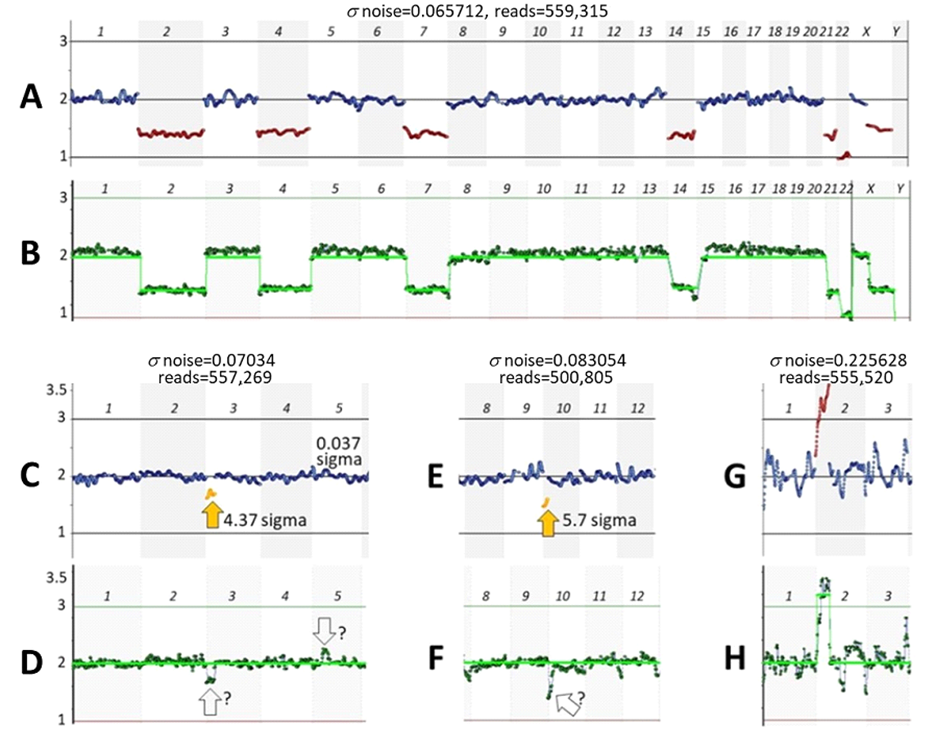
Fig. 2: Simulating a desired noise level. A sample was obtained using the VeriSeq PGS kit and analyzed with CNVector (A, B, and C) or BlueFise Multi (D) software. A-Copy number calls for each chromosome; B, C, and D-CNV charts (only chromosomes 1–8 are shown). The noise level (s = 0.06746) (A, B) of this sample was adjusted to a noise level that is the median for the VeriSeq PGS kit (s = 0.099937) (C) by reducing the number of reads from 604,189 (A, B) to 151,681 (C). A BlueFuse Multi CNV chart of the sample with an unreduced number of reads is shown for comparison (D). Arrows point to segmental trisomy on chromosome 1 and segmental monosomy on chromosome 7 with assigned N sigma values.
From a low-noise sample (s = 0.06746) that has segmental p36.33–p31.1 trisomy in chromosome 1 and segmental monosomy p22.3–p21.1 in chromosome 7 (Fig. 2A, 2B), the noise was increased by 0.0325 sigma (Fig. 2C), which is close to the difference between the noise medians characteristic of each kit (Fig. 1). Note that with increased s noise (Fig. 2C), the statistical significance of detecting mosaicisms (N sigma) was reduced, though in this case it still remained high. The analysis of the same sample using CNVector software (Fig. 2B) was in concordance with the analysis using BlueFuse Multi software (Fig. 2D).
Detection of mosaicisms, especially small segmental mosaicisms, is an excellent approach for rigorous characterization of PGT-A methods, as these are more difficult to detect than whole chromosome monosomies or trisomies. In such studies, mosaicisms were often created by mixing aneuploid and euploid cell lines in different proportions, thus simulating different levels of mosaicism [4, 5, 25]. This study also simulated mosaicisms but used more precise mixing in silico (Fig. 3).
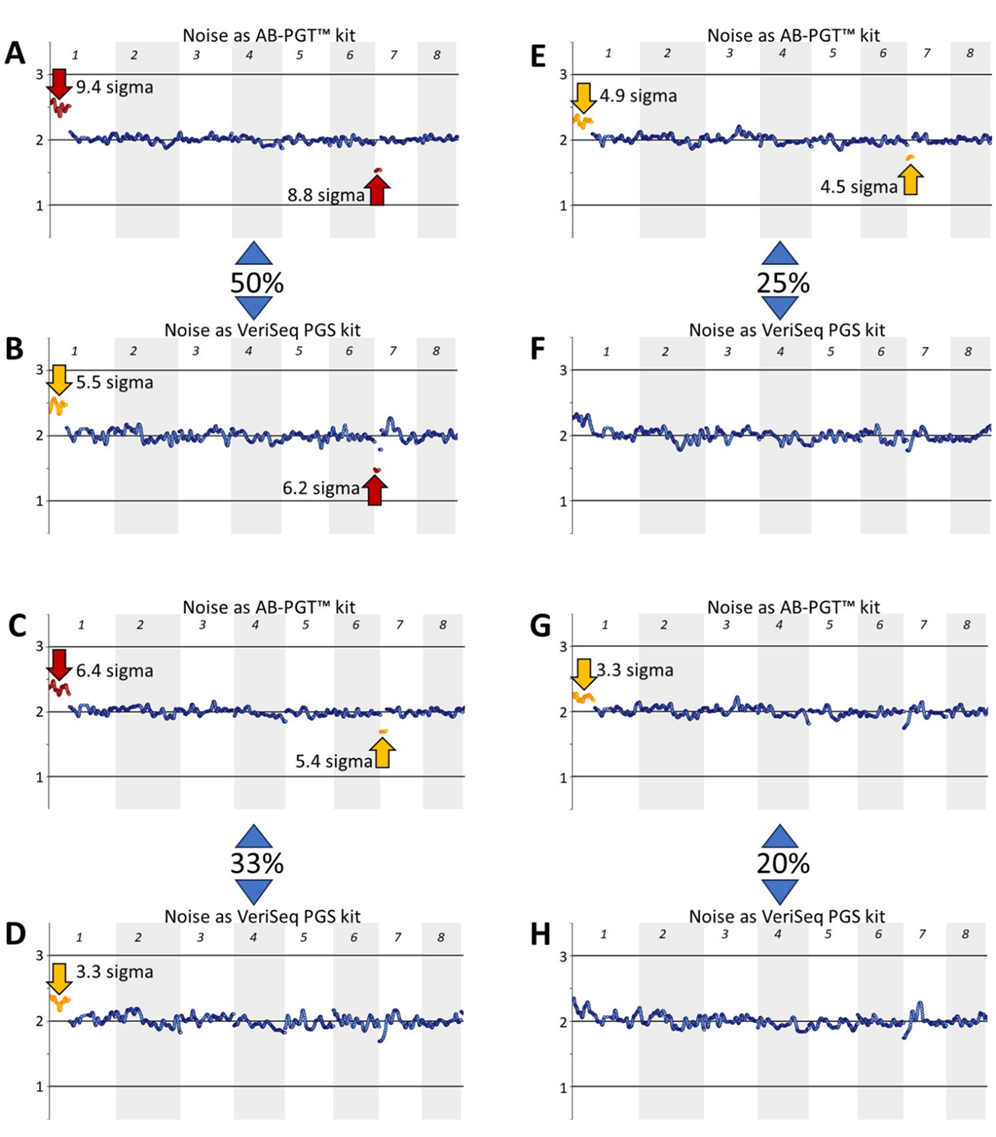
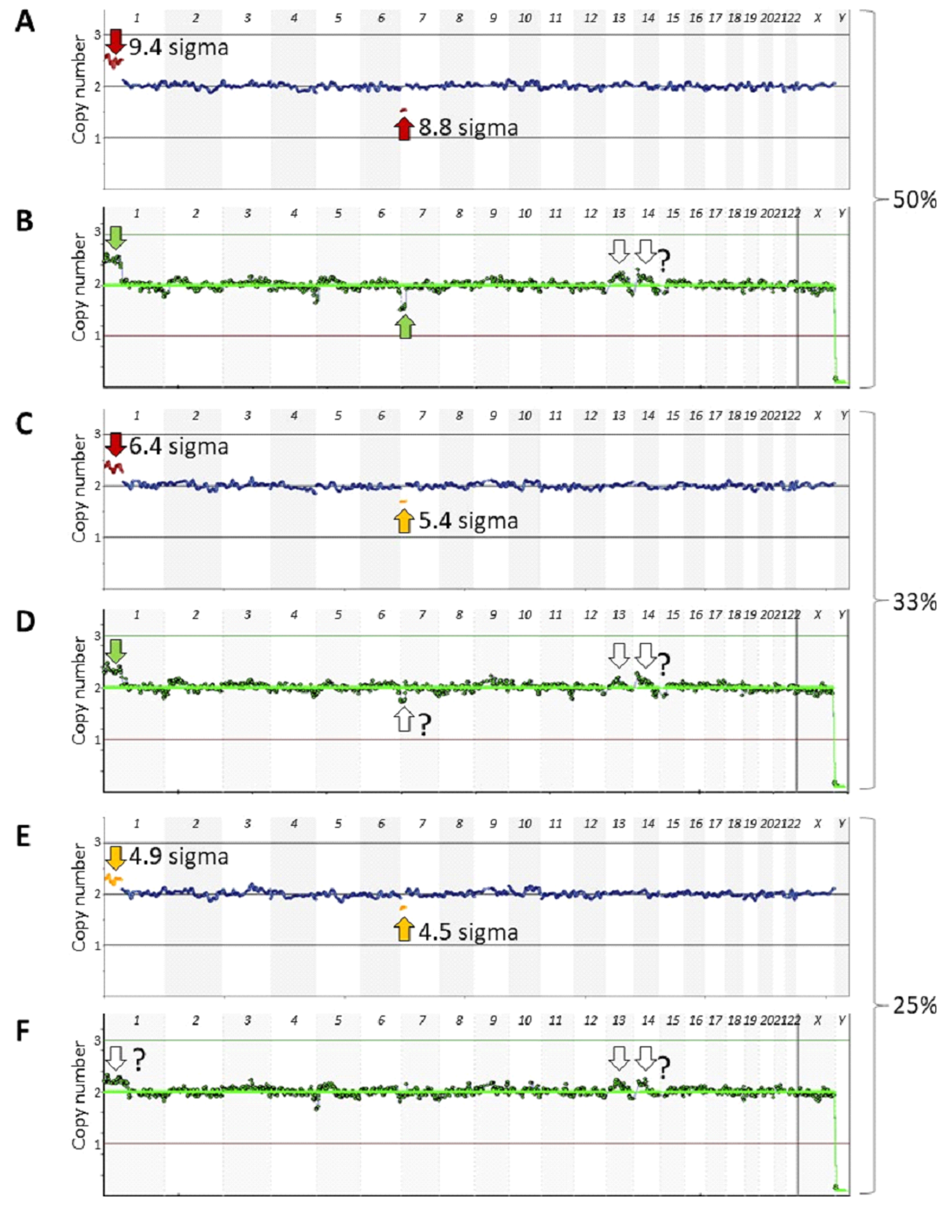
Fig. 3: Detection of 50%, 33%, 25%, and 20% mosaicisms at about median noise levels characteristic of either the AB-PGT or VeriSeq PGS kits. Euploid and aneuploid samples were obtained using the VeriSeq PGS kit, and their noise levels were adjusted to about the median noise levels characteristic of either the AB-PGT (s ∼ 0.07) or VeriSeq PGS (s ∼ 0.1) kits. Reads of the adjusted samples with the same noise level were mixed at 50% (A, B), 33% (C, D), 25% (E, F), and 20% (G, H) of the aneuploid sample and analyzed using CNVector software. Only chromosomes 1–8 are shown on CNV charts. Numerals 3, 2, and 1 at the left of the charts indicate copy number positions.
To this end, noise levels in euploid and aneuploid samples were adjusted to median levels characteristic for AB-PGT (s ∼ 0.07) or VeriSeq PGS (s ∼ 0.1) kits at about 500,000 reads, the number common for both methods. Different read numbers could have been used, as Ds noise between the medians does not depend on the number of reads (Fig. 1). Next, reads from euploid and aneuploid samples of the same noise level were randomly mixed at 50%, 33%, 25%, and 20% of the aneuploid sample, thus simulating corresponding levels of mosaicism. The statistical significance of mosaicism detection progressively decreased with a decreasing percentage of the aneuploid sample (Fig. 3). At the 50% mosaicism level, both mosaicisms were detected at both noise levels (Figs. 3A, 3B). However, at the 33% mosaicism level, the segmental loss in chromosome 7 was detected at about the AB-PGT noise level (Fig. 3C), but it was missed at the VeriSeq PGS noise level (Fig. 3D). At the 25% mosaicism level, both aberrations were still detected at the AB-PGT noise level (Fig. 3E), whereas they were not detected at the VeriSeq PGS noise level (Fig. 3F). The 20% segmental loss in chromosome 7 was not detected at both noise levels (Figs. 3G, 3H).
Comparison of CNVector and BlueFuse Multi software
Full karyotype concordance was observed when analyzing VeriSeq PGS euploid samples and samples with monosomies and trisomies, e.g., (Figs. 2B, 2D, 4A, 4B), as well as samples with ∼ 55% mosaicisms (Figs. 4A, 4B) using either CNVector or BlueFuse Multi software.
However, the analysis a lower-level mosaicisms and mosaicisms at the flanks of the chromosomes was challenging using the latter as it involves arbitrary visual assessment. For example, in Fig. 4D, there appear to be low-level mosaicisms in chromosomes 3 and 5. Analysis of the same sample using CNVector software (Fig. 4C) confirms the former mosaicism (4.37 sigma) but excludes the latter because of practically no deviation from the norm in chromosome 5 (0.037 sigma). CNVector software detected about 50% segmental loss in chromosome 10 (Fig. 4E). However, it was not flagged by the BlueFuse Multi perhaps because, unlike with CNVector, artifacts at the flanks of the chromosomes are common with the former (e.g., at the ends of chromosomes 11 and 12 in Fig. 4F). Interestingly, trisomies and monosomies were often reliably detected even at very high noise levels (Figs. 4G, 4H). In these cases, N sigma has a value of approximately 1/s, and hence their detection is often confident even when noise levels are high. However, this is not the case for mosaicisms where N sigma = DCN/s, where 0 < DCN < 1 is the CN deviation from normality of the mosaic region. As DCN is reduced, it becomes increasingly difficult to detect a mosaicism with a high level of statistical significance. Accordingly, operating with noise levels above s = 0.125 should be avoided, as less prominent aberrations could be missed.
We also investigated performance of both software at different mosaicism levels (Fig. 5). The data set shown in Fig. 3 for AB-PGT noise level was used for the comparison. At the 50% mosaicism level, both aberrations were apparent with either software (Figs. 5A, 5B), and 33% and 25% mosaicisms were also detected using CNVector software (Figs. 5C, 5E). However, at the 33% level, a smaller aberration in chromosome 7 was hardly visible using BlueFuse Multi (Fig. 5D) and was completely undetectable at the 25% level (Fig. 4F), consistent with the recommendation that BlueFuse Multi is not designed to automatically detect mosaicisms below a 50% level (A Technical Guide to Aneuploidy Calling with VeriSeq PGS, Part # 15059470).
Discussion
In this study, we compared the performance of two kits using a novel metric—the median noise level assessed on 973 routine clinical samples. Reduced overall noise in AB-PGT samples leads to a threefold improvement in multiplexing samples for sequencing as compared to VeriSeq PGS samples. In order to objectively compare the entire AB Vector and Illumina workflows, we also had to compare their software. A comparison of BlueFuse Multi and CNVector software demonstrated better performance by the latter on less than 50% mosaicisms and comparable performance on larger aberrations, thus further supporting this assessment. Importantly, multiplexing more samples per run brings down the cost per sample, which is a major expense in in vitro fertilization (IVF) [26, 27]. So far, the expectation that the reduced costs of NGS would lead to a fall in the PGT-A price per sample [7, 28] has not been realized. NGS methods offered considerable improvements in the quality of PGT-A analysis in comparison with aCGH [29, 30], resulting in better clinical outcomes [31]. The performance of aCGH could be limited by the non-specific hybridization of probes [32]. However, NGS methods are impeded by the variable number of reads, that are, unlike aCGH probes, not in fixed positions throughout the genome. This results in Poisson noise (Fig. 1), which is a major impediment in NGS-based PGT-A methods. The Poisson noise is random, meaning it is uncorrelated between neighboring bins in the CNV analysis. This contrasts with the systematic noise and bias that are caused by certain biochemical events, such as a polymerase that is stalled on GC-rich sequences in PCR.
The main difference between the AB Vector and Illumina kits is the proprietary SuperDOP WGA technology, which is an optimized degenerate oligonucleotide-primed PCR (DOP-PCR) [33]. With the DOP-PCR, a single WGA reaction rather than two is required, thus simplifying manipulations and minimizing the risk of contamination. However, DOP-PCR is inefficient compared to modern methods and suffers from low genome coverage [12, 34, 35]. In SuperDOP, this limitation was overcome by optimization for improved priming on the template, resulting in an overall decrease in the number of PCR cycles required. While in DOP-PCR, 5 LS cycles are followed by 25–35 HS cycles [33], SuperDOP uses 2 LS cycles followed by 12 HS cycles, resulting in only 14 PCR cycles in total. In the SurePlex kit, 12 LS pre-amplification cycles are followed by 14 HS amplification cycles, resulting in a total of 26 cycles, or nearly twice as many as used for WGA in the AB-PGT kit. Minimizing the overall number of PCR cycles is desirable, as systematic bias against extremely GC-rich and AT-rich sequences accumulates with each cycle [36]. Minimizing the number of LS cycles is particularly important. During multiple LS cycles, amplicons that by chance were generated at initial cycles can serve as templates at subsequent LS cycles, resulting in overrepresentation of corresponding areas of the genome (bias).
Besides, PGT-A performance can be compromised by a low-quality sequencing run. For example, instead of an optimal 500,000 reads passing filter, 250,000 reads were observed after re-sequencing of the same sample [8]. With fewer reads, the CNV profile became less clear (more noisy), compromising the detection of a segmental aneuploidy, which agrees with our data (Figs. 1, 3). Other possible sources of noise are imperfections in the equipment and variabilities in the quality control of the kit components. The latter is contingent upon the complexity of the kits, with variability increasing with more components and more complex steps.
With the intention of attaining the best resolution, sensitivity studies are typically performed on a few arbitrarily selected samples of good quality. This is useful for measuring the detection threshold with different methods on such samples [5, 10]. However, it provides little information on the overall performance of a method in a clinical setting as it relates to a small number of choice samples. For example, low-noise samples are obtained using either Illumina or AB Vector kits. However, the best 15% of AB Vector samples have s < 0.0625, whereas only 2% of Illumina samples presented in Fig. 1 satisfy this condition. Conversely, 24% of the worst Illumina samples have s > 0.125, whereas only 3.6% of AB Vector samples exhibit such high noise. Another shortcoming of current methods is that sensitivity would be by far superior if an aberration resides in a genome area densely covered with reads, as opposed to an aberration that resides in a sparsely covered area. Thus, it is difficult to compare different studies claiming certain resolution of a method in megabases, unless they used the same aberration and the quality of the samples (noise level) was the same, which is hardly ever the case. Therefore, commonly used “sensitivity” and “limit of detection” parameters are of limited value as they pertain to only a few high-quality samples. On the contrary, the median noise level parameter is characteristic of an overall method’s performance in a clinical setting and includes all samples, regardless of their quality or ploidy status. Unlike sensitivity and specificity, the noise can be automatically, unambiguously, and quantitatively measured, as in this study.
We observed large variations in noise levels (Fig. 1), while even modest differences affected the detection of mosaicisms (Fig. 3). Therefore, concordance studies such as those with blastocyst rebiopsy samples [37–40] could benefit from normalization of noise, as in this study. Nevertheless, currently there are no guidelines on how to deal with technical noise in mosaicism studies [2]. Implantation failure after euploid blastocyst transfer ranges from 25% to 50% [41, 42], which could be in part due to undetected mosaicisms [5, 29, 30], especially in “noisy” samples. It has been noted that it is “crucial to define if and to what extent a poor-quality biopsy, rather than a pure biological issue, may result in a plot suggestive of mosaicism” [43]. We observed vastly different noise levels in different samples regardless of sample preparation method, which seems like a common but underreported phenomenon. Others have also observed large differences in the quality of biopsy samples, which are suggested to be due to operator error [38]. The reasons for the large variations in noise levels of biopsy samples obtained by a skilled operator remain unclear. They may be associated with patients' age, with different morphokinetic categories of embryos [44, 45], or with metabolic signatures [46]. Biopsies may contain broken cells that were damaged by the biopsy needle and were not removed by a wash; the zona pellicular that contains cell debris [47] may be biopsied along with trophectoderm cells, all contributing to the noise. According to data from different fertility centers, biopsy techniques affect the pregnancy outcome, and suboptimal biopsy techniques could cause damage to embryos [2, 3, 48–50]. Blastocysts that are not particularly high in quality are especially susceptible to damage [40], and there is no standardization of blastocyst biopsy procedures between the centers [20], again increasing variability. There is no standardized system to interpret and report PGT-A data either [2]. We suggest that, besides standardizing PGT-A methods, the median noise parameter could also serve as a common quantitative indicator of practices in fertility centers. Since success rates in different centers widely vary [41, 42], this robust parameter could be instrumental in improving the centers’ practices either via self-regulation or with the involvement of a government agency [51].
Additional Information
A.B. declares that he holds shares and is an employee of AB Vector, LLC (www.abvector.com). He does not have any other unpaid roles or relationships that might have a bearing on the publication process. M.T., S.P., I.M., and D.F. declare no conflict of interest.
Acknowledgements
The authors are grateful for constructive criticism and improvement of the manuscript to Professor Ian Jones, Department of Biological Sciences, University of Reading, UK; Anugraha Raman, Computational Biologist at the University of California San Diego, San Diego, USA; Vladimir Kaimonov, Director, Genetico Genetics Laboratory, Moscow, Russia; and Viktor Volkomorov, Senior Scientist, Genomics LLC, Tbilisi, Georgia.
Abbreviations
In Vitro Fertilization (IVF); Preimplantation Genetic Testing for Aneuploidies (PGT-A); Copy Number (CN); Copy Number Variation (CNV); Next Generation Sequencing (NGS); array Comparative Genomic Hybridization (aCGH); Degenerate Oligonucleotide-Primed PCR (DOP-PCR); Whole Genome Amplification (WGA); Low-stringency (LS); High-stringency (HS); Standard Deviation (SD).
References
- Hassold T, Hunt, P. To err (meiotically) is human: The genesis of human aneuploidy. Nat Rev Genet 2 (2001): 280–291.
- Viotti M. Preimplantation genetic testing for chromosomal abnormalities: aneuploidy, mosaicism, and structural rearrangements. Genes (Basel) 11 (2020): 602.
- Chen HF, Chen M, Ho HN. An overview of the current and emerging platforms for preimplantation genetic testing for aneuploidies (PGT-A) in in vitro fertilization programs. Taiwan J Obstet Gynecol 59 (2020): 489-495.
- Chuang TH, et al. High concordance in preimplantation genetic testing for aneuploidy between automatic identification via Ion S5 and manual identification via Miseq. Sci Rep 23 (2021): 18931.
- Biricik A, et al. Cross-validation of next-generation sequencing technologies for diagnosis of chromosomal mosaicism and segmental aneuploidies in preimplantation embryos model. Life (Basel) 11(2021): 4:340.
- Gutierrez-Mateo C, et al. Validation of microarray comparative genomic hybridization for comprehensive chromosome analysis of embryos. Fertil Steril 95 (2011): 953-958.
- Zheng H, Jin H, Liu L, et al. Application of next-generation sequencing for 24-chromosome aneuploidy screening of human preimplantation embryos. Mol Cytogenet 8 (2015): 38.
- Cuman C, et al. Defining the limits of detection for chromosome rearrangements in the preimplantation embryo using next generation sequencing. Hum Reprod 33 (2018): 1566-1576.
- Li N, et al. The performance of whole genome amplification methods and next-generation sequencing for pre-implantation genetic diagnosis of chromosomal abnormalities. J Genet Genomics 42 (2015): 151-159.
- Deleye L, et al. Performance of four modern whole genome amplification methods for copy number variant detection in single cells. Sci Rep 7 (2017): 3422.
- Volozonoka L, Miskova A, Gailite L. Whole genome amplification in preimplantation genetic testing in the era of massively parallel sequencing. Int J Mol Sci 23 (2022): 4819.
- Arneson N, Hughes S, Houlston R, et al. Whole-genome amplification by degenerate oligonucleotide primed PCR (DOP-PCR). CSH Protoc 4919 (2008).
- Popov S, et al. Validation of a new technology for whole genome amplification (WGA) and NGS sequencing in preimplantation genetic testing (PGT). ESHG (2020).
- Trevethan R. Sensitivity, specificity, and predictive values: foundations, pliabilities, and pitfalls in research and practice. Front Public Health 5 (2017): 307.
- Fordham DE, et al. Embryologist agreement when assessing blastocyst implantation probability: is data-driven prediction the solution to embryo assessment subjectivity? Hum Reprod 37 (2022): 2275-2290.
- Neves AR, Montoya-Botero P, Polyzos NP. The role of androgen supplementation in women with diminished ovarian reserve: time to randomize, not meta-analyze. Front Endocrinol 12 (2021): 653857.
- Patounakis G, Hill Complexities and potential pitfalls of clinical study design and data analysis in assisted reproduction. Curr Opin Obstet Gynecol 30 (2018): 139-144.
- Gleicher N, Kushnir VA, Barad DH. How PGS/PGT-A laboratories succeeded in losing all credibility. Reproductive biomedicine online 37 (2018): 242-245.
- Grati FR, et al. Response: how PGS/PGT-A laboratories succeeded in losing all credibility. Reprod Biomed Online 37 (2018): 246.
- Munné S, Yarnal S, Martinez-Ortiz PA, et al. Response: how PGS/PGT-A laboratories succeeded in losing all credibility. Reprod Biomed Online 37 (2018): 247-249.
- Doyle N, et al. Donor oocyte recipients do not benefit from preimplantation genetic testing for aneuploidy to improve pregnancy outcomes. Hum Reprod 35 (2020): 2548-2555.
- Pos O, et al. DNA copy number variation: main characteristics, evolutionary significance, and pathological aspects. Biomed J 44 (2021): 548-559.
- Franklin A. Prologue: the rise of the sigmas. In: Shifting standards. Experiments in Particle Physics in the Twentieth Century. University of Pittsburgh Press, Pittsburgh PA 15260 (2013).
- Varian H. Bootstrap tutorial. Mathematica Journal 9 (2005): 768-775.
- Treff NR, et al. Validation of concurrent preimplantation genetic testing for polygenic and monogenic disorders, structural rearrangements, and whole and segmental chromosome aneuploidy with a single universal platform. Eur J Med Genet 62 (2019): 103647.
- Theobald R, Sengupta S, Harper J. The status of preimplantation genetic testing in the UK and USA. Hum Reprod 35 (2020): 986-998.
- Somigliana E. Cost-effectiveness of preimplantation genetic testing for aneuploidies. Fertil Steril 111 (2019): 1169-1176.
- Yang Z, et al. Randomized comparison of next-generation sequencing and array comparative genomic hybridization for preimplantation genetic screening: a pilot study. BMC Med Genomics 8 (2015): 30.
- Munné S, Grifo J, Wells D. Mosaicism: "survival of the fittest" versus "no embryo left behind" Fertil Steril. 105 (2016): 1146-1149.
- Maxwell SM, et al. Why do euploid embryos miscarry? A case-control study comparing the rate of aneuploidy within presumed euploid embryos that resulted in miscarriage or live birth using next-generation sequencing. Fertil Steril 106 (2016): 1414-1419.
- Munné S, et al. Clinical outcomes after the transfer of blastocysts characterized as mosaic by high resolution Next Generation Sequencing- further insights. Eur J Med Genet 63 (2020): 103741.
- Tan DSP, Lambros MBK, Natrajan R, et al. Getting it right: designing microarray (and not ‘microawry’) comparative genomic hybridization studies for cancer research. Laboratory Investigation 87 (2007): 737-754.
- Telenius H, et al. Degenerate oligonucleotide-primed PCR: general amplification of target DNA by a single degenerate primer. Genomics 13 (1992): 718-25.
- Navin NE. Cancer genomics: one cell at a time. Genome Biol 15 (2014): 452.
- Huang L, Ma F, Chapman A, et al. Single-cell whole-genome amplification and sequencing: methodology and applications. Annu Rev Genomics Hum Genet 16 (2015): 79-102.
- Aird D, et al. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol 12 (2011): 2.
- Victor AR, et al. Assessment of aneuploidy concordance between clinical trophectoderm biopsy and blastocyst. Hum Reprod 34 (2019): 81-192.
- Sachdev ND, McCulloh DH, Kramer Y, et al. The reproducibility of trophectoderm biopsies in euploid, aneuploid, and mosaic embryos using independently verified next-generation sequencing (NGS): a pilot study. J Assist Reprod Genet 37 (2020): 559-571.
- Marin D, Xu J, Treff NR. Preimplantation genetic testing for aneuploidy: A review of published blastocyst reanalysis concordance data. Prenat Diagn 41 (2021): 545-553.
- Mizobe Y, et al. The effects of differences in trophectoderm biopsy techniques and the number of cells collected for biopsy on next-generation sequencing results. Reprod Med Biol 21 (2022): e12463.
- Mastenbroek S, Twisk M, Van der Veen F, et al. Preimplantation genetic screening: a systematic review and metaanalysis of RCTs. Hum Reprod 17 (2011): 454-466.
- Mastenbroek S, Repping S. Preimplantation genetic screening: back to the future. Hum. Reprod 29 (2014): 1846-1850.
- Cimadomo D, et al. The impact of biopsy on human embryo developmental potential during preimplantation genetic diagnosis. Biomed Res Int 2016 (2016): 7193075.
- Gardner DK, Schoolcraft WB. In vitro culture of human blastocysts. In: Jansen R., Mortimer D., editors. Towards Reproductive Certainty: Fertility and Genetics Beyond. Parthenon Publishing; Nashville, TN, USA: (1999): 378–388.
- Basile N, et al. The use of morphokinetics as a predictor of implantation: A multicentric study to define and validate an algorithm for embryo selection. Hum Reprod.30 (2015): 276-283.
- Pallisco R, et al. Metabolic signature of energy metabolism alterations and excess nitric oxide production in culture media correlate with low human embryo quality and unsuccessful pregnancy. Int J Mol Sci 24 (2023): 890.
- Aizer A, Harel-Inbar N, Shan H, et al. Can expelled cells/debris from a developing embryo be used for PGT? J Ovarian Res 14 (2021): 104.
- Cimadomo D, et al. Inconclusive chromosomal assessment after blastocyst biopsy: prevalence, causative factors and outcomes after re-biopsy and re-vitrification. A multicenter experience. Hum Reprod 33 (2018): 1839-1846.
- Rubino P, et al. Trophectoderm biopsy protocols can affect clinical outcomes: time to focus on the blastocyst biopsy technique. Fertil Steril113 (2020): 981-989.
- Makhijani R, et al. Impact of trophectoderm biopsy on obstetric and perinatal outcomes following frozen–thawed embryo transfer cycles. Hum Reprod36 (2021): 340-348.
- Yang H, DeWan AT, Desai MM, et al. Preimplantation genetic testing for aneuploidy: challenges in clinical practice. Hum. Genomics 16 (2022): 69.


 Impact Factor: * 5.3
Impact Factor: * 5.3 Acceptance Rate: 75.63%
Acceptance Rate: 75.63%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 