Telomere Dynamics, Gene Expression and Genetic Instability in Glioblastoma Cells Treated with Reversine
Fábio Morato de Oliveira1*, Aline Monezi Montel1, Wagner Gouvêa dos Santos1, Fermino Sanches Lizarte Neto2, Cristina Mores Junta3, Ubirajara Lanza Júnior4, Sabine Mai5
1Laboratory of Medical Genetics, Federal University of Jataí, Brazil
2Faculty of Medicine of Ribeirão Preto, University of São Paulo, Brazil
3Faculty of Santa Casa de Belo Horizonte - Ensino e Pesquisa. Belo Horizonte, Minas Gerais, Brazil
4Department of Pharmacy-University Center of Votuporanga, São Paulo, Brazil
5CancerCare Manitoba Research Institute, CancerCare Manitoba, The Genomic Centre for Cancer Research and Diagnosis, The University of Manitoba, Winnipeg, Canada, MB R3E 0V9, Canada
*Corresponding author: Fábio Morato de Oliveira, Laboratory of Medical Genetics, Federal University of Jataí, Brazil.
Received: 27 September 2023; Accepted: 06 October 2023; Published: 06 November 2023
Article Information
Citation: Fábio Morato de Oliveira, Aline Monezi Montel, Wagner Gouvêa dos Santos, Fermino Sanches Neto, Cristina Mores Junta, Ubirajara Lanza Júnior, Sabine Mai. Telomere Dynamics, Gene Expression and Genetic Instability in Glioblastoma Cells Treated with Reversine. Journal of Biotechnology and Biomedicine. 6 (2023): 565-572.
DOI: 10.26502/jbb.2642-91280118
View / Download Pdf Share at FacebookAbstract
Background: In the present study, we analyzed the cytotoxic effect of reversine, a small molecule used for stem cell dedifferentiation and potential to selectively induce cell death, on two human glioblastoma cell lines. The AURKA and AURKB gene expression were quantified in both cell lines following the exposure to different concentrations of reversine, and the effect of reversine on telomere dynamics, in a 3D scale was analyzed. AURKA and AURKB genes express mitotic kinases with an important role in the regulation of several mitotic events. Hyperexpression of these genes are found in patients with cytogenetic abnormalities presenting an unfavorable prognosis.
Methods and Results: Our results indicate that reversine was able to inhibit the cell growth of both cell lines in a time and concentration-dependent fashion. A comparative analysis showed that reversine induced apoptosis of U138 cells more significantly than in U87 cells at the same dosage. Reversine was able to decrease the expression levels of AURKA and AURKB mRNA in both cell lines after 24 hours exposure. The total number of telomeres in both glioblastoma cells was determined after 48 h exposure and compared (not treated vs. treated cells) using TeloView®™ software. Two distinct subgroups of the same cell line were identified, based on treatment status (reversine+/-). Microscopy Fluorescence analysis showed glioblastoma cells exhibiting nuclei with low relative fluorescent intensities suggestive of short telomeres and nuclei with intermediate relative fluorescent intensity indicating intermediate length of telomeres.
Conclusions: Our results have demonstrated that reversine can act as an important antiproliferative agent in glioblastoma cells and possibly promote redifferentiation of glioblastoma cell lines by genetic reprogramming and changing the genomic instability status, as suggested by decreased gene expression profile, and two distinct telomere signatures. Aurora kinase inhibitors such as reversine may have potential therapeutic value when combined with radiation therapy.
Keywords
<p>Glioblastoma; Reversine; Telomere; Aurora Kinase; Genomic instability</p>
Article Details
Introduction
Glioblastoma represents the most common malignant primary brain tumor, with an incidence rate of 3.2 cases per 100,000 in the United States. It is characterized by a high proliferation index, cellular heterogeneity, and highly capacity to infiltrate adjacent normal brain tissue [1]. Genomic instability, characterized by chromosomal abnormalities, telomere disfunction and differential gene expression are very commom in glioblastoma, occurring in 80 to 85% of adult patients [2]. Despite notorious poor survival rates, patients with glioblastoma exhibit a sequence of outcomes ranging from a survival time of several weeks to several years [3]. Telomeres are complex structures of DNA and proteins, at the ends of chromosomes that play an important role in cellular maintenance [4]. On the other hand, telomere dysfunction has been associated with predisposition to chromosomal rearrangements, genomic instability and in final analysis malignant cell transformation [5]. From a three-dimensional nuclear perspective, in normal cells telomeres are homogeneously organized and localized in micro territories [6]. In contrast, telomeres of tumor cell nuclei show an altered three-dimensional nuclear organization with manifestation of telomeric aggregates [7]. Alterations in the nuclear telomere architecture and telomeric dysfunction are associated with chromosomal instability (structural and numerical), which is a hallmark of glioblastoma cells [8]. Regarding the genomic instability, aurora kinase genes (AURKA and AURKB) play a critical role in mitosis by regulating centrosome duplication, bipolar spindle formation, alignment of chromosomes on the mitotic spindle, and the mitotic checkpoint [9]. High expression of aurora kinase genes has been implicated in many types of neoplasia including breast, gastric, colon, ovarian, liver, non–small cell lung, uterine, esophageal and leukemias [10,11]. In CNS tumors, overexpression of aurora kinase genes in medulloblastoma, low-grade gliomas and glioblastoma, in addition to high Ki-67 expression has been reported [12-15]. Reversine (2-(4-Morpholinoanilino)-6-cyclohexylaminopurine) is a small synthetic molecule that promotes in vitro and in vivo differentiation of human fibroblasts into skeletal muscle cells [16,17]. It has been identified as a novel class of ATP-competitive aurora kinase inhibitor acting through the formation of the reversine-Aurora kinase complex. Reversine can reprogram somatic cells to a state of increased plasticity that can be manipulated to direct differentiation in various cell types. In addition, this small molecule displays an anti-cancer activity in some cancer cell lines, such as multiple myeloma, leukemia cells, thyroid cancers, oral squamous carcinoma cells among other tumor cell types [18-21]. Therefore, in the present study, we analyzed the cytotoxic role of reversine on two glioblastoma cell lines (U87 and U138). Furthermore, we analyzed the AURKA and AURKB expression in both cell lines under exposure to different concentrations of reversine, and determined the effect of reversine on telomere dynamics, in a 3D scale.
Material and Methods
Cell lines and cell culture and Reversine treatment
The human glioblastoma cell lines U87 and U138 were obtained from the American Type Culture Collection (ATCC). The cells were cultivated in Eagle’s minimum essential medium (Thermo Fisher Scientific, Waltham - Massachusetts, EUA), supplemented with 15% of fetal bovine serum (Thermo Fisher Scientific, Waltham - Massachusetts, EUA) and 1% of penicillin/streptomycin (Thermo Fisher Scientific, Waltham - Massachusetts, EUA). The cells were maintained at 37°C, with 5% of CO2. Reversine was purchased from Sigma-Aldrich (USA), prepared in dimethyl sulfoxide (DMSO), in accordance with the manufacture’s instruction. The cells were plated onto 24-well tissue-culture plates and grown (2.0x105 cells/well) in medium. After twelve hours, the cells were treated with pure medium (containing 0.01% DMSO) as control and medium containing different concentrations of reversine (5.0, 10.0, 15.0, 20.0, 50.0, 100.0, 150.0 and 200.0nM). The cells were incubated for 24h, 48h and 72h.
Cell viability analysis.
For cell viability analysis we performed methylthiazoletetrazolium (MTT) assays and trypan blue exclusion testing. For MTT, 5000 glioblastoma cells were seeded/well in a 96-well plate. After 24h, the cells were treated with different concentrations of Reversine (5.0, 10.0, 15.0, 20.0, 50.0, 100.0, 150.0 and 200.0nM). DMSO was used as a negative control. Cell viability was tested by the MTT assay for 24, 48 and 72h. MTT solution was added to each well and incubated at 37°C for 4h. The absorbance was finally determined at 490 nm using a microplate reader. For the trypan blue test, 1.0 x 104 glioblastoma cells from each cell line were seeded/well on 24-well plates. After a 24h the cells were treated with different concentrations of reversine (5.0, 10.0, 15.0, 20.0, 50.0, 100.0, 150.0 and 200.0nM). Following a 24-72h incubation period, cell viability was assessed by counting live versus dead cells on a hemocytometer using the Trypan Blue (0.4% in phosphate-buffer saline, PBS) exclusion staining. The test was performed in triplicate. Results were expressed using GraphPad Prism 8 (GraphPad Software, Inc., San Diego, CA, USA).
Aurora kinase mRNA analysis.
RNA was isolated from glioblastoma cells treated with reversine, using TRIzol reagent (Thermo Fisher Scientific, Waltham - Massachusetts, EUA), as previously described [22]. Complementary DNA (cDNA) was synthesized from ~1μg of total RNA using a High-Capacity cDNA reverse transcription Kit (Thermo Fisher Scientific, Waltham - Massachusetts, EUA), following the manufacturer's instructions. For analysis of aurora kinase genes, primes and probe developed by Assay on Demand were used (AURKA: Hs00269212_m1 and AURKB: Hs00177782_m1; Thermo Fisher Scientific, Waltham - Massachusetts, EUA). The AURKA and AURKB genes and GAPDH mRNA, used as endogenous internal control for each sample, were analyzed in duplicate on the same MicroAmp optical 96-well plates using a 7500 Real-Time PCR System (Thermo Fisher Scientific, Waltham - Massachusetts, EUA). Real-time quantitative polymerase chain reaction (RT-qPCR) assays were performed in a final reaction volume of 20μl. The comparative cycle threshold (Ct) method was used to determine the relative expression level of AURKA and AURKB genes. On comparative analysis of glioblastoma samples and healthy donors, AURKA and AURKB gene expression was calculated as a relative quantification to the GAPDH housekeeping gene. The gene expression AURKA and AURKB from glioblastoma samples was calculated as relative quantification to healthy donors (ΔΔCt = ΔCt patient – ΔCt healthy donors+) and expressed as 2−ΔΔCt. All primers were standardized by conventional semi-quantitative PCR analysis before proceeding to the real-time quantitative analysis.
3-D Quantitative Fluorescent in Situ Hybridization (Q-FISH).
For Q-FISH analysis, slides containing fixed glioblastoma cells were incubated in 3.7% formaldehyde/1xPBS solution for 10 minutes, followed by incubation in 20% glycerol/1xPBS solution for 45min. The cells were treated by four repeated cycles of freeze-thaw in glycerol. Then, the slides were incubated in 0.1 HCL solution and fixation was performed in 70% formamide/2xSCC for 1 hour. For hybridization, slides were covered with 8μl of PNA telomeric probe (Agilent Dako, Santa Clara, California, USA), sealed with coverslip and rubber cement. For denaturation, the slides were placed on a hot plate, protected from direct light, for 3 minutes, at 82°C. The hybridization was carried out for 2 hours, at 30°C. The slides were then washed three times in 70% formamide/10mM Tris (pH 7.4) solution, for 15 minutes followed by washing in 1xPBS at room temperature for 2 minutes, and in 0.1xSSC at 55°C for 5 minutes under shaking. Finally, the slides were washed in 2xSSC/ 0.05% Tween 20 solution for three times, for 5 minutes, at room temperature under shaking. After final washing, the nuclei were counter-stained with 4’,6-diamino-2-phenylindole (DAPI) (0.1μg/ml) and antifade reagent (Thermo Fisher Scientific, Waltham - Massachusetts, EUA), and covered with coverslip for image acquisition.
3D image acquisition and analysis using TeloView®™ software.
30 interphase nuclei were analyzed, for each sample, by using an AxioImager M1 microscope (Carl Zeiss, Jena, Germany), coupled to an AxioCam HRm charge-coupled device (Carl Zeiss, Jena, Germany) and a 63-x oil objective lens (Carl Zeiss, Jena, Germany). The acquisition time was 500 milliseconds (ms) for Cy3 (telomeres) and 5 ms for DAPI (nuclei). Sixty z-stacks were acquired at a sampling distance of x,y: 102 nm and z: 200 nm for each slice of the stack. AxioVision 4.8 software (Carl Zeiss, Jena, Germany) was used for 3D image acquisition. Deconvolved images were converted into TIFF files and exported for 3D-analysis using the TeloView®™ software (Telo Genomics Corp., Toronto, ON, Canada) [22].
Data image analysis – 3D telomere architecture
The evaluation of the telomeric architecture of glioblastoma cells was performed by TeloView®™ software. It measures five distinct parameters for each sample: (1) the number of telomeres (signals); (2) the total intensity (telomere size); (3) the distribution and frequency of telomere aggregates, which means clusters of telomeres that are found in proximity that cannot be further resolved; (4) the a/c ratios represent a measure defined by the cell cycle progression through interphase cells. Thus, it is possible to check if there was a difference in cell cycle among the glioblastoma samples. The telomere dynamics through varies with cell cycle; according to stages of the cell cycle (G0/G1, S, and G2); (5) Finally, TeloView®™ software allows us to measure the distance of each telomere signal from the nuclear center versus the periphery. For the distinct subgroups of glioblastoma cells, a graphical representation was obtained showing the distribution of the intensity of the acquired telomere fluorescent signals, the distribution of the frequency of telomere aggregates per cell and the acquired signals per cell.
Statistical analysis for differential expression analysis (AURKA/AURKB)
All data obtained are presented as the mean ± standard deviation. GraphPad Prism 8.0 software (Graph Pad Software, La Jolla, CA, USA) was used for statistical analysis. Statistical analysis was performed with one-way ANOVA of.
Statistical Analysis for telomere architecture
Based on treatment of glioblastoma cells with reversine, two distinct subgroups were defined. The telomeric parameters (number, length, telomere aggregates, nuclear volumes, and a/c ratio) were compared between these subgroups using analysis of variance. All telomere parameters in both glioblastoma subgroups were compared using chi-square analysis. On the other hand, cell parameters averages were analyzed using nested factorial analysis of variance. Significance level was set at 0.05.
Results
To determine the effect of reversine, we measured the cell proliferation rate in two human glioblastoma cell lines (U87 and U138) after exposure. Reversine was able to inhibit the cell growth of both cell lines in a time and concentration-dependent way (Figure1A-D). We also defined the half maximal inhibitory concentration (IC50) for both cell lines. Using a nonlinear regression analysis, the IC50 obtained were 89.1nM(24h), 34.1nM(48h) and 33.3nM(72h) for U87 and 85.0nM(24h), 59.7nM(48h) and 22.3nM(72h) for U138. We observed that the viable reversine-treated cells changed morphologically from a swollen shape with less cell-to-cell connections to a homogeneous appearance. The cell number decreased with reversine treatment, showing the anti-proliferative effect of reversine on both cell lines. A comparative analysis evidenced that the potency of reversine to induce cell apoptosis was more significant in U-187 cells than in U87 cells at the same dosage. After determination of cell cytotoxicity by reversine treatment, we evaluated the expression levels of AURKA and AURKB mRNA, by exposing glioblastoma cells to different concentrations of reversine. Our results showed that reversine was also a potent inhibitor of AURKA and AURKB gene expressions. Both protein-products are required for mitotic chromosome segregation, spindle checkpoint function, cytokinesis, and histone H3 phosphorylation. Our experiments demonstrated that the levels of AURKA and AURKB mRNA were downregulated in both glioblastoma cell lines (Figure 2A-B). The 24h exposure of reversine, allowed to verify that the expression levels of AURKA and AURKB mRNA decreased by 55.2% and 38.1%, respectively, in the U87 cell line. In contrast, for U138 cell line, the AURKA expression levels declined more than AURKB; 59.4% and 36.8%, respectively. Futhermore, reversine was proven to block proliferation or to induce programmed cell death in different malignant cell lines, this small molecule, can induce dedifferentiation and cell reprogramming. Various studies have focused on the roles of reversine in regenerative medicine15,16. In this context, telomere dynamics analysis has documented that it was possible to distinguish among cell transformation, from a “higher to a lower level” of genomic instability, by the frequency of telomere aggregates, through treatment with reversine. For telomere analysis we only considered the IC50 for both cell lines, and 48h of reversine exposure. We analyzed the total number of telomeres in both glioblastoma cells (not treated vs. treated cells) using TeloView®™ software22 and thereafter observed three categories of samples (Figures 3 and 4). In figure 3, it is shown telomeric nuclear signals of each glioblastoma cell line (U87 and U138) not treated (first and second column and reversine treated, third column) in two-dimensional and three-dimensional images using reconstruction after deconvolution. Forty nuclei per cell line and treatment status were identified in blinded analysis. TeloView®™ indicated the presence of two distinct subgroups of the same cell line, based on treatment status (reversine +/-). We also observed nuclei with low relative fluorescent intensities representing glioblastoma cells with short telomeres and nuclei with intermediate relative fluorescent intensity indicating intermediate length of telomeres (Figure 4). The telomere organization inside the nuclei varies according to cell cycle. Thus, the determination of the a/c ration represent a way to define progression through cell cycle in interphase cells. It is necessary to keep in mind that the nuclear space that contains the telomeres is defined by two axes, a and b, that are equal in length, and a third axis, c, that has a different length22. In our study we find significant differences for the a/c ratio (p < 0.05) and for nuclear volume (p < 0.05) among the two groups (reversine +/-), for both glioblastoma cell lines.
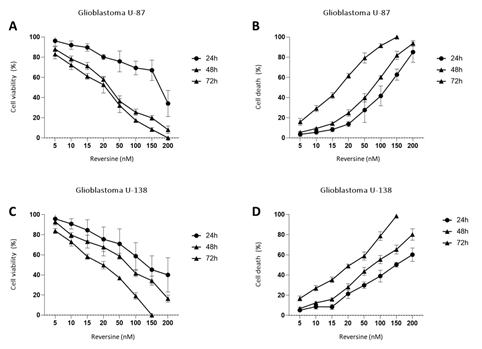
Figure 1: Reversine inhibited the proliferation of glioblastoma cells cells. U-87 and U-138 glioblastoma cells were treated with different concentrations of Reversine (5, 10, 15, 20, 50, 100, 150 and 200 nM) for 24, 48 and 72 hours. Results represent the mean ±SD of four experiments each performed in triplicate. A/B - Cell viability was determined by MTT assay for U-87 and U-138 cells treated with reversine. Values were normalized with DMSO treated cells. C/D - Dose- and time-response cytotoxicity curves analyzed by trypan blue assay for glioblastoma cells treated with reversine. Values were expressed as percentage of viable cells for each condition. Results are shown as mean±SD of three independent experiments; p<0.0001; ANOVA test.
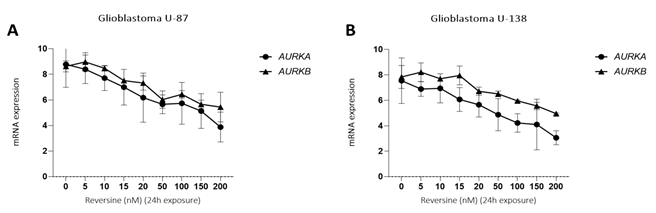
Figure 2: qPCR analysis of AURKA and AURKB mRNA expression in glioblastoma cells treated with reversine (5, 10, 15, 20, 50, 100, 150 and 200 nM) for 24h DMSO solution was used as calibrator sample. The graph represents the mean±SD of three independent experiments. The p values are indicated in the graphs; *p<0.05, **p<0.01; ANOVA test and Bonferroni post-test.
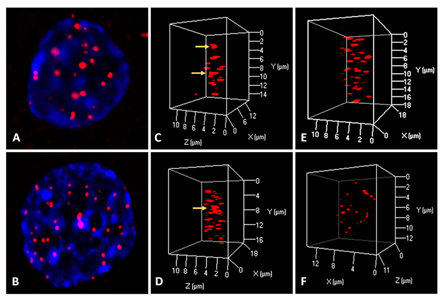
Figure 3: Representative 2D (A, B) and 3D (C, D, E, F) telomere analysis in glioblastoma cells using a PNA telomeric probe. Glioblastoma cell U-87 not treated (C), and U-87 cell treated with reversine (34.1nM) for 48h. The yellow arrows identify the presence of telomere aggregates. For U-138 glioblastoma cell telomere aggregates were observed in cell without treatment (D), and not seen in U-138 cell treated with Reversine (59.7nM) for 48h.
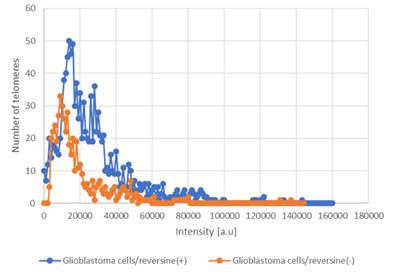
Figure 4: Graph distribution of number of telomeres according to their intensity (length of telomeres) for glioblastoma cells [treated with reversine/not treated - reversine (+/-)]. The image represents the 3D telomere distribution of the 3D telomeric profile.
Discussion
Reversine was discovered in 2004, by Chen and co-workers as a small molecule able to induce myogenic lineage-committed cells into multipotent progenitor cells [23]. A couple of years later, 24. Hsieh and co-workers (2007) [24], observed that reversine was capable to inhibits cell proliferation of various human cancer cells. The relationship among reversine and aurora kinase genes became evident after Anna Morena and colleagues [18] have demonstrated that reversine could induce failure in cytokinesis and subsequent polyploidy in cells. Considering its lower toxicity on normal cells compared to others aurora kinase inhibitors, reversine has entered in phase II clinical trials [25,26]. In our investigation, we tested the effects of reversine over glioblastoma cell lines in order not only to confirm its antiproliferative/cytotoxic effects, but also to verify the possible effect of reversine on the 3D telomere dynamics. The treatment with reversine induced nuclear condensation and fragmentation, a hallmark of apoptosis, in both glioblastoma cell lines. Reversine effectively inhibited the growth of U87 and U138 cells in a concentration dose dependent. This observation is in accordance with results previously published, regarding the anticancer effects of reversine over several types of cancer cells [18-21,24-26]. We also observed the overexpression of aurora kinase genes in both glioblastoma cell lines and the expanded analysis demonstrating reduction of the expression levels of AURKA and AURKB, after exposure to reversine. In 24h, reversine was able to reduce the mRNA levels of AURKA and AURKB in 55.2% and 38.1%, respectively, in the U87 cell line. In contrast, for U138 cell line, the AURKA expression levels declined more than AURKB; 59.4% and 36.8%, respectively. According to previous studies, aurora kinase mRNA present itself significantly upregulated in glioblastoma compared with normal brain specimens and pilocytic astrocytoma’s. In addition, high AURKA mRNA expression was significantly associated with higher grade CNS neoplasms, and this characteristic may represent a general marker of tumor malignancy [27,28]. In gliomas, a high expression of AURKA or AURKB is associated with a malignant phenotype and a poor prognosis and chemoresistance [29-31]. Telomeres are essential DNA sequences responsible to form a stable and recognized end of the chromosome that avoids it being perceived from the cell as a naked chromosome. In terms of nuclear organization, the telomeres present a particular disposition within the three-dimensional space of the nuclei, in which telomeres from normal cells do not overlap, and are localized in micro territories [6,32]. Modifications to this nuclear organization leads to the initiation of genomic instability [33]. Studies using high-resolution fluorescence imaging techniques in combination with quantitative optical analyses, indicated that the nuclei harbors chromosomes that are nonrandomly positioned within the 3D nuclear space [6,34,35]. On the other hand, telomeres from neoplastic cells present themselves as an irregular nuclear organization with the presence of telomeric aggregates [7]. In this investigation, the spatial telomere organization was obtained using the TeloView® software. TeloView® analysis was able to determine the subclassification of telomere organization regarding the reversine treatment status of the cells. Thus, our investigation identified two distinct populations of glioblastoma cells based on the number of telomere aggregates, telomere number, and size of telomeres. Studies regarding telomere dynamics in glioblastoma revealed for the first time that three glioblastoma subgroups could be distinguished according to their nuclear telomeric patterns, and these profiles corresponded to patient survival and disease progression [36,37]. Keeping in mind the variability of patient responses to treatment, the clinical application of telomere signatures could represent a powerful prognostic biomarker in glioblastoma. In summary, we have demonstrated that reversine can act as an important antiproliferative agent against glioblastoma cells and promote redifferentiation of glioblastoma cell lines from a “high” to a “lower” genomic instability status, as demostrated by two distinct telomere signatures. The three-dimensional telomere analysis confirmed that the number and distribution of telomeres differed markedly between the glioblastoma cells treated and not treated with reverisne. TeloView®™ analysis of the sample slides displays telomere numbers, sizes, and aggregates. It is possible that in the future we could use this classification strategy to identify patients whose Aurora kinase inhibitors use could be beneficial for tumor shrinkage. A rational approach would be to enrich early trials with patients whose aurora kinase expression is greater than the mean expression of patients with glioblastoma. Additional studies are needed to explore this approach and to determine if it is more suitable to measure aurora kinase expression at the gene or protein level in patient specimens. The present study suggests that aurora kinase inhibitors may be of greater therapeutic value when combined with radiation therapy due to the observed synergy.
Compliance with Ethical Standards
This is an experimental study involving commercial tumor cell lines. The Research Ethics Committee has confirmed that no ethical approval is required.
Acknowledgments
The authors would like to thank the Telo Genomics Corp., Toronto, ON, Canada for the use of TeloView®™. The authors also thank the Genomic Centre for Cancer Research and Diagnosis (GCCRD) for imaging. The GCCRD is funded by the Canada Foundation for Innovation and supported by CancerCare Manitoba Foundation, the University of Manitoba, and the Canada Research Chair Tier 1 (S.M.). The GCCRD is a member of the Canadian National Scientific Platforms (CNSP) and of Canada BioImaging.
Conflicts of Interest
S.M. is a shareholder, director, and chair of the clinical and scientific advisory board of Telo Genomics Corp. (Toronto, ON, Canada). The other authors declare that they have no conflicts of interest.
Funding
The study was funded by the Canadian Institutes of Health Research (SM)
Authors' contributions
FMO: Have been involved in drafting the manuscript and revising it critically for important intellectual content; WGS: Responsible for the provision and analysis of samples; FSN: Responsible for the provision and analysis of samples; CMJ: Revised the entire work and add important intellectual content; ULJ: Revised the entire work and add important intellectual content; SM: Supervised the entire work, revised it critically for important intellectual content. All authors have given final approval of the version to be published. Each author has participated sufficiently in the work to take public responsibility for appropriate portions of the content.
References
- Louis, DN, Ohgaki H, Wiestler OD, et al. World Health Organization Histological Classification of Tumors of the Central Nervous System; International Agency for Research on Cancer: Lyon, France (2016).
- Malhotra A, Lindberg M, Faust GG, et al. Breakpoint profiling of 64 cancer genomes reveals numerous complex rearrangements spanned by homology-independent mechanisms. Genome Res 23 (2013): 762-776.
- Wang J, Cazzato E, Ladewig E, et al. Clonal evolution of glioblastoma under therapy. Nat Genet 48 (2016) 768-776.
- De Lange T. How telomeres solve the end-protection problem. Science 326 (2009): 948-952.
- Hemann MT, Strong MA, Hao LY, et al. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell 107 (2001): 67-77.
- Chuang TC, Moshir S, Garini Y, et al. The three-dimensional organization of telomeres in the nucleus of mammalian cells. BMC Biol 2 (2004): 12.
- Mai S, Garini Y. The significance of telomeric aggregates in the interphase nuclei of tumor cells. J Cell Biochem 97 (2006): 904-915.
- Gadji M, Fortin D, Tsanaclis AM, et al. Three-dimensional nuclear telomere architecture is associated with differential time to progression and overall survival in glioblastoma patients. Neoplasia 12 (2010): 183-191.
- Giet R, Petretti C, Prigent C. Aurora kinases, aneuploidy and cancer, a coincidence or a real link? Trends Cell Biol 15 (2005): 241-250.
- Kitzen JJ, de Jonge MJ, Verweij J. Aurora kinase inhibitors. Crit Rev Oncol Hematol 73 (2010): 99-110.
- Yan M, Wang C, He B, et al. Aurora-A Kinase: A Potent Oncogene and Target for Cancer Therapy. Med Res Rev 36 (2016): 1036-1079.
- Klein A, Reichardt W, Jung V, et al. Overexpression and amplification of STK15 in human gliomas. Int J Oncol 25 (2004): 1789-1794.
- Neben K, Korshunov A, Benner A, et al. Microarray-based screening for molecular markers in medulloblastoma revealed STK15 as independent predictor for survival. Cancer Res 64 (2004): 3103-3111.
- Reichardt W, Jung V, Brunner C, et al. The putative serine/threonine kinase gene STK15 on chromosome 20q13.2 is amplified in human gliomas. Oncol Rep 10 (2003): 1275-1279.
- Samaras V, Stamatelli A, Samaras E, et al. Comparative immunohistochemical analysis of aurora-A and aurora-B expression in human glioblastomas. Associations with proliferative activity and clinicopathological features. Pathol Res Pract 205 (2009): 765-773.
- Lee EK, Bae GU, You JS, et al. Reversine increases the plasticity of lineage-committed cells toward neuroectodermal lineage. J Biol Chem 284 (2009): 2891-2901.
- Anastasia L, Sampaolesi M, Papini N, et al. Reversine-treated fibroblasts acquire myogenic competence in vitro and in regenerating skeletal muscle. Cell Death Differ 13 (2006): 2042-2051.
- D'Alise AM, Amabile G, Iovino M, et al. Reversine, a novel Aurora kinases inhibitor, inhibits colony formation of human acute myeloid leukemia cells. Mol Cancer Ther 7 (2008): 1140-1149.
- Lu CH, Liu YW, Hua SC, et al. Autophagy induction of reversine on human follicular thyroid cancer cells. Biomed Pharmacother 66 (2012): 642-647.
- Lee YR, Wu WC, Ji WT, et al. Reversine suppresses oral squamous cell carcinoma via cell cycle arrest and concomitantly apoptosis and autophagy. J Biomed Sci 19 (2012): 9.
- Kuo CH, Lu YC, Tseng YS, et al. Reversine induces cell cycle arrest, polyploidy, and apoptosis in human breast cancer cells. Breast Cancer 21 (2014): 358-369.
- Oliveira FM, Lucena-Araujo AR, Favarin Mdo C, et al. Differential expression of AURKA and AURKB genes in bone marrow stromal mesenchymal cells of myelodysplastic syndrome: correlation with G-banding analysis and FISH. Exp Hematol 41 (2013): 198-208.
- Chen S, Zhang Q, Wu X, et al. Dedifferentiation of lineage-committed cells by a small molecule. J Am Chem Soc 126 (2004): 410-411.
- Hsieh TC, Traganos F, Darzynkiewicz Z, et al. The 2,6-disubstituted purine reversine induces growth arrest and polyploidy in human cancer cells. Int J Oncol 31(2007): 1293-1300.
- McMillin DW, Delmore J, Weisberg E, et al. Tumor cell-specific bioluminescence platform to identify stroma-induced changes to anticancer drug activity. Nat Med 16 (2010): 483-489.
- Qin HX, Yang J, Cui HK, et al. Synergistic antitumor activity of reversine combined with aspirin in cervical carcinoma in vitro and in vivo. Cytotechnology 65 (2013): 643-653.
- Lehman NL, O'Donnell JP, Whiteley LJ, et al. Aurora A is differentially expressed in gliomas, is associated with patient survival in glioblastoma and is a potential chemotherapeutic target in gliomas. Cell Cycle 11 (2012): 489-502.
- Goeppert B, Schmidt CR, Geiselhart L, et al. Differential expression of the tumor suppressor A-kinase anchor protein 12 in human diffuse and pilocytic astrocytomas is regulated by promoter methylation. J Neuropathol Exp Neurol 72 (2013): 933-941.
- Reichardt W, Jung V, Brunner C, et al. The putative serine/threonine kinase gene STK15 on chromosome 20q13.2 is amplified in human gliomas. Oncol Rep 10 (2003): 1275-1279.
- Pattwell SS, Konnick EQ, Liu YJ, et al. Neurotrophic Receptor Tyrosine Kinase 2 (NTRK2) Alterations in Low-Grade Gliomas: Report of a Novel Gene Fusion Partner in a Pilocytic Astrocytoma and Review of the Literature. Case Rep Pathol 2020 (2020): 5903863.
- Reichardt W, Jung V, Brunner C, et al. The putative serine/threonine kinase gene STK15 on chromosome 20q13.2 is amplified in human gliomas. Oncol Rep 10 (2003): 1275-1279.
- Vermolen BJ, Garini Y, Mai S, et al. Characterizing the three-dimensional organization of telomeres. Cytometry A 67 (2005): 144-150.
- Gadji M, Vallente R, Klewes L, et al. Nuclear Remodeling as a Mechanism for Genomic Instability in Cancer. In: Gisselsson D., editor. Advances in Cancer Research. Volume 112. Academic Press; Oxford, UK (2011): 77-126.
- Song C, Takagi M, Park J, et al. Analytic 3D imaging of mammalian nuclei at nanoscale using coherent x-rays and optical fluorescence microscopy. Biophys J 107 (2014): 1074-1081.
- Kempfer R, Pombo A. Methods for mapping 3D chromosome architecture. Nat Rev Genet 21 (2020): 207-226.
- Gadji M, Fortin D, Tsanaclis AM, et al. Three-dimensional nuclear telomere architecture is associated with differential time to progression and overall survival in glioblastoma patients. Neoplasia 12 (2010): 183-191.
- Gadji M, Mathur S, Bélanger B, et al. Three-Dimensional Nuclear Telomere Profiling as a Biomarker for Recurrence in Oligodendrogliomas: A Pilot Study. Int J Mol Sci 21 (2020): 8539.


 Impact Factor: * 5.3
Impact Factor: * 5.3 Acceptance Rate: 75.63%
Acceptance Rate: 75.63%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 