MTNR1A Promoter Methylation is Associated with Increased Breast Cancer Risk
Johanna Samulin Erdem1, Øivind Skare1, Marte Petersen-Øverleir1, Heidi Ødegaard Notø1, Jenny-Anne S. Lie1, Kristina Kjærheim2, Edyta Reszka3, Beata Peplonska4, Shanbeh Zienolddiny1*
1National Institute of Occupational Health, Oslo, Norway
2Cancer Registry of Norway, Oslo, Norwayx
3Department of Molecular Genetics and Epigenetics, Nofer Institute of Occupational Medicine, Lodz, Poland
4Department of Environmental Epidemiology, Nofer Institute of Occupational Medicine, Lodz, Poland
*Corresponding Author: Dr. Shanbeh Zienolddiny, National Institute of Occupational Health, PO Box 8149 Dep, N-0033 Oslo, Norway
Received: 24 June 2020; Accepted: 17 July 2020; Published: 03 August 2020
Supplementary File
Article Information
Citation: Samulin Erdem J, Øivind Skare, Marte Petersen-Øverleir, Heidi Ødegaard Notø, Jenny-Anne S. Lie, Kristina Kjærheim, Edyta Reszka, Beata Pepłońska, Shanbeh Zienolddiny. MTNR1A Promoter Methylation is Associated with Increased Breast Cancer Risk. Journal of Cancer Science and Clinical Therapeutics 4 (2020): 245-255.
View / Download Pdf Share at FacebookAbstract
Shift work, particularly, night work has been classified as a probable carcinogen to humans based on the increased risk observed in epidemiological studies for some cancer types, including female breast cancer. The underlying molecular mechanisms are not well established, but may involve aberrant epigenetic modifications. Here, effects of changes in methylation status of 5-methyl cytosine in melatonin and female hormone receptor genes were investigated as possible mechanisms for increased breast cancer risk in female night shift workers. Methylation in promoter regions of the MTNR1A, MTNR1B, PGR, ESR1 and ESR2 genes was analyzed by pyrosequencing in a nested case-control study of female nurses, including 354 breast cancer cases and 356 healthy controls. The effects of methylation as well as the combined effects of methylation and shift work on breast cancer risk were assessed. We demonstrate that increased methylation of the MTNR1A promoter is associated with increased risk of breast cancer (OR=1.13, 95% CI: 1.02 -1.24, P=0.019). No association between promoter methylation levels and breast cancer risk was observed for the other receptor genes investigated. Furthermore, MTNR1A methylation levels were not affected by shift work. Altogether, our data suggest that epigenetic regulation of MTNR1A may contribute to breast cancer.
Keywords
<p>DNA methylation; Night shift; Breast cancer; Melatonin receptor</p>
Article Details
Abbreviation:
5mC: 5-methyl cytosine; CTCF: CCCTC-binding factor; ESR1: estrogen receptor alpha; ESR2: estrogen receptor beta; EZH2: enhancer of zeste homolog 2; FSH: follicle stimulating hormone; GCF: GC-rich sequence DNA-binding factor; LAN: light at night; LH: luteinizing hormone; OR: odds ratio; MI: methylation index; MTNR1A: melatonin receptor 1A; MTNR1B: melatonin receptor 1B; PGR: progesterone receptor, RAD21:RAD21 cohesion complex component; TP53: tumor protein p53
1. Introduction
Breast cancer is the most commonly diagnosed cancer and the leading cause of cancer death in women. The etiology of breast cancer is complex and may involve multiple biological, lifestyle and genetic risk factors [1-5]. Approximately, 30% of breast cancer incidence can be attributed to established risk factors such as young age at menarche, late age at menopause, late age at first birth, nulliparity, hormone replacement therapy, exposure to ionizing radiation, and alcohol consumption [2, 5]. Occupational risk factors may also contribute to breast cancer risk [6, 7]. Based on several epidemiological studies, night work is probably one of the occupational risk factors that may increase risk of breast cancer [8-18], and night shift work has been classified as probably carcinogenic (Group 2A) by IARC in 2019 [19]. A study on Nordic female cabin crew members found an increased risk (SIR, 1.5) for breast cancer [12]. Similarly, Danish female military staff had an odds ratio of 3.9 for breast cancer in women exposed to intense night shifts of longer duration and morning chronotype [10]. Stratifying the intensity of night work exposure by number of consecutive night shifts, a case control study of Norwegian nurses showed significantly increased risk of breast cancer (OR 1.7–1.8) in nurses that worked five or more years in work schedules including more than six consecutive night shifts, compared with nurses who never worked night shifts [9]. A similar Danish study also found an increased risk of breast cancer in nurses working night shift with various shift schedules (OR 2.9) [11]. A more recent European pooled population based case-control study also confirmed that cumulative rotating night shift work is associated with increased breast cancer risk [14]. While most studies indicate that night shift work is significantly associated with higher breast cancer risk, other studies have provided insignificant evidence for this relationship [20-23]. However, a recent meta-analysis identifying a positive association between night shift work and breast cancer, concludes that cancer risk increases with cumulative years of night shift work [15].
Although most of the epidemiological studies are suggestive of an association between shift work and breast cancer, the mechanisms are not well studied. Shift work may involve disruption of several biological pathways including the disruption of metabolic and biological circadian clock. Shift work, implying exposure to light at night (LAN) and the subsequent disruption of the synthesis of melatonin, has been suggested as a potential biological mechanism causing disruption of the circadian rhythmicity [24, 25]. Accordingly, LAN exposure leads to decreased melatonin levels that are important for regulation of circadian rhythmicity, as melatonin regulates the transcriptional activity of circadian and circadian targeted genes [26]. Melatonin acts through its membrane bound receptors melatonin receptor 1A and 1B, which are encoded by the MTNR1A and MTNR1B genes. Melatonin may also act through binding to intracellular proteins, such as calmodulin, or through binding to orphan nuclear receptors. In addition to melatonin’s role in the regulation of the circadian clock, a role of melatonin in the regulation of apoptosis has also been suggested [27], although, not all studies have confirmed this [28]. Finally, melatonin may influence steroid hormone levels by affecting protein levels of gonadotropin releasing hormone and thereby the secretion of follicle stimulating hormone (FSH), and luteinizing hormone (LH) that controls the activity of the gonads [29]. Moreover, melatonin may modulate the activity of the aromatase enzyme, converting androgens to estrogens, i.e. androstendione to estrone and testosterone to estradiol [30]. The steroid hormones estrogen and progesterone are known risk factors and play important roles in breast tumorigenesis by regulating breast epithelial cell proliferation, differentiation, and apoptosis. The action of estrogen is regulated by its receptors, estrogen receptor alpha (ERα) and beta (ERβ), which are encoded by the ESR1 and ESR2 genes, respectively. Similarly, the effect of progesterone is regulated by the two major receptor protein isoforms PGRA and PGRB, which are encoded by a single progesterone receptor (PGR) gene. Estrogen and progesterone levels define the hormone-responsive phenotypes of breast tumors [31], allowing classifications of breast cancer subgroups by ESR and PGR status, i.e. being positive or negative for these receptors. Interestingly, melatonin suppresses both ESR1 expression and estrogen-induced transcriptional activity of ESR1 in ER-positive human breast cancer cells [32], and night shift has been found to be associated with ESR1 hypomethylation [33]. Furthermore, melatonin may also regulate the transcriptional activity of other enzymes involved in estrogen metabolism [26]. Understanding the biological mechanisms behind development of shift work-related breast cancer may aid in evidence-based regulatory policymaking. The present case-control study aimed to address the hypothesis that night work might influence cancer risk through epigenetic mechanisms, namely through 5-methyl cytosine (5mC) in female steroid and melatonin hormonal pathways. To achieve this, our analyses focused on two melatonin (MTNR1A and MTNR1B) and three steroid hormone (PGR, ESR1and ESR2) receptor genes. The 5mC in the proximal promoter regions of these genes were mapped using pyrosequencing in DNA from breast cancer cases and matched controls stratified by shift work parameters.
2. Materials and methods
2.1 Study population
Approval for the study was obtained from the Regional Committee for Medical and Health Research Ethics, South-East region (S-08430a, 2008/10453). All participants signed an informed consent document. Study subjects were participants of a larger cohort of 49,402 female nurses graduated between 1914 and 1985. The design of the nested case-control study has been described in detail previously [9] and is outlined in Supplementary Figure S1. All participants in the study had worked as nurses for at least one year. Cases were diagnosed with breast cancer between 1990 and 2007 at an age of 35-74 years and alive by February 2009. Controls were frequency matched to cases in five-year age groups by diagnostic year of the case. Only controls that were cancer-free at and prior to the year of diagnosis of the case were included. All participants were interviewed by telephone to obtain information on potential breast cancer risk factors and lifetime occupational history. Prior to the interviews, an information letter containing a checklist for work history, a letter of consent, a request for saliva samples, and an Oragene saliva sampling kit (DNA Genotek Inc, Kanata, ON, Canada) was sent to the participants. A total of 563 cases and 619 controls were included in the study based on the availability of obtained saliva samples and information of past history of shift work.
2.2 Exposure assessment
Information of night shift work was obtained by telephone interviews conducted in 2009. Work history including information on job duration, workplace, proportion of fulltime work and work schedules (only day, only night, or both day and night shifts) were collected. Night shift was defined as a shift including work between 12 pm and 6 am. Night work included working periods from both rotating and permanent night schedules. For each night shift working period, information on number of consecutive night shifts (intensity) was obtained. For more detailed information of the exposure assessments the reader is referred to our previous publication [9]. Exposure metrics based on a combination of duration and intensity of night work were used in this study. The utilized exposure metric involved the intensity of night work (i.e. more or less than three consecutive night shifts) with duration of night work (i.e. more or less for less than five years). Accordingly, participants were grouped into four groups based on their night work exposure patterns: no exposure (day workers, never), low exposure (< three consecutive night shifts), medium exposure (≥ three consecutive night shifts < five years) and high exposure (≥ three consecutive night shifts ≥ five years).
2.3 Methylation analysis
A random subset of cases (n=354) and controls (n=356) was included in the epigenetic analysis of 5mC. Cases and controls were matched according to the work schedules across the defined night work exposure groups. Methylation of MTNR1A, MTNR1B, PGR, ESR1, and ESR2 promoters were analyzed by pyrosequencing using Pyromark Q24 Advanced technology (Qiagen). Pyrosequencing primers were designed using the PyroMark Assay Design software (Qiagen), Supplementary Table S1. Primers were located to functional regions based on transcription factor binding sites utilizing ENCODE project database (www.encode.org) and the PROMO software in the TRANSFAC database (http://gene-regulation.com/pub/databases.html) [34], Supplementary Figure S2 and Supplementary Table S2. For each gene the number of analyzed CpG dinucleotides was as follows: MTNR1A=12, MTNR1B=6, PGR=9, ESR1=7 and ESR2=4. In short, DNA from saliva samples was extracted using Oragene DNA isolation kit as described by the manufacturer (DNA Genotek Inc.). DNA samples were bisulfite treated using EpiTect Fast Bisulfite Conversion kit (Qiagen, Hilden, Germany) and PCR amplified using unbiased nested primers and the PyroMark PCR kit (Qiagen) according to the manufacturer’s instructions. The percentage methylation for each of the target CpG sites for each of the respective genes was calculated using the PyroMark CpG Software (Qiagen). A methylation index (MI) was calculated for each CpG site as the mean percentage of methylation across all analyzed CpG dinucleotides.
2.4 Statistical analyses
Characteristics of the study subjects were assessed by Chi-square or Mann-Whitney U-test as appropriate in IBM SPSS software version 23.0. Differences in MI between cases and controls, and between different exposure categories were analyzed for each gene. Odds ratios of breast cancer were analyzed using logistic regression. The data was ln-transformed prior to analysis to ensure normal distribution of the data. Separate analyses were performed for 1) cancer status and 2) interaction between night work exposure and cancer status. In all analyses, the list of potential confounders tested included: alcohol consumption, parity, duration of daily occupational exposure to x-rays, hormonal treatment during the last two years before diagnosis, age at saliva test, years since cancer diagnosis, and occurrence of familiar breast cancer. All possible combinations of adjustment variables were compared and the combination that minimized the AIC was chosen. Final correction variables are listed in the footnote of each table. In each analysis crossed random intercepts were included for subject and CpG island to take into account the repeated observations for the CpG islands. All presented data were transformed back to the original scale. P≤0.05 was considered statistically significant.
3. Results
The characteristics of the study subjects and the exposure variables of night shift work are shown in Supplementary Table S3. As expected, known risk factors such as a higher occurrence of familial breast cancer (P=0.001), and a lower number of children (P=0.033) among cases were significantly different from the controls. Moreover, alcohol consumption was higher in cases (9.5%) than in controls (5.3%), (P=0.052). No significant difference was observed for use of hormone replacement therapy the last two years before diagnosis, between cases and controls. MIs for each of the five genes were generally low in all subjects regardless of case- control status and the average MI ranged from two to eight percent with the highest mean MI in the MTNR1B promoter (8.38%) and the lowest MI (2.41%) in the ESR1 gene, Figure 1.
Analysis of effects of receptor methylation status on breast cancer occurrence showed that increased levels of MTNR1A MI were associated with increased risk of breast cancer (OR=1.13 95% CI: 1.02 – 1.24, P=0.019), Table 1. Methylation status of estrogen and progesterone receptors was not associated with breast cancer risk. Detailed analysis of individual CpGs in the MTNR1A gene showed that methylation of predominately CpG 1 was associated with increased breast cancer risk (OR=1.12, 95% CI: 1.04 – 1.20, P=0.002), Table 2. Assessment of effects of shift work schedules on the methylation pattern of MTNR1A in CpG 1 in cases and controls showed that shift work did not considerably affect MTNR1A CpG 1 methylation, Table 3.
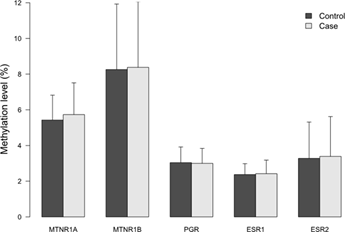
Figure 1: Methylation indices (MIs) of the five receptor genes in cases and controls. Results represent mean methylation ± SD.
|
Gene |
OR (95% CI)a |
P |
|
MTNR1A |
1.13 (1.02 – 1.24) |
0.019 |
|
MTNR1B |
1.01 (0.96 – 1.05) |
0.800 |
|
PGR |
0.96 (0.80 – 1.14) |
0.621 |
|
ESR1 |
1.08 (0.85 – 1.39) |
0.524 |
|
ESR2 |
1.03 (0.95 – 1.10) |
0.489 |
aOR was analyzed using logistic regression. Methylation index was averaged over all CpGs. Adjustments were made for parity, alcohol consumption and familiar breast cancer. Significant p-values ≤ 0.05 are indicated in bold.
Table 1: Odds ratios for breast cancer according to an 1-unit increase in methylation index (M1) on breast cancer.
|
CpG # |
OR (95% CI)a |
P |
|
1 |
1.12 (1.04 – 1.20) |
0.002 |
|
2 |
1.08 (1.00 – 1.16) |
0.048 |
|
3 |
1.01 (0.98 – 1.05) |
0.490 |
|
4 |
1.06 (0.98 – 1.13) |
0.131 |
|
5 |
1.02 (0.98 – 1.06) |
0.386 |
|
6 |
1.02 (0.95 – 1.09) |
0.611 |
|
7 |
1.03 (0.97 – 1.09) |
0.387 |
|
8 |
1.04 (1.00 – 1.08) |
0.071 |
|
9 |
1.03 (0.97 – 1.09) |
0.389 |
|
10 |
1.04 (0.99 – 1.10) |
0.133 |
|
11 |
1.04 (0.98 – 1.10) |
0.233 |
|
12 |
0.98 (0.91 – 1.06) |
0.678 |
aOR was analyzed using logistic regression. Adjustments were made for parity, alcohol consumption and familiar breast cancer. Significant p-values ≤ 0.05 are indicated in bold.
Table 2: Odds ratios for breast cancer according to an 1-unit increase in methylation index (MI) at individual CpGs in the MTNR1A gene.
|
Exposure |
N1 |
Controls |
P1 |
N2 |
Cases |
P2 |
Cases vs controls Estimate (95% CI) |
P3 |
|
never nights |
88 |
5.19 (4.79,5.62) |
REF |
72 |
5.68 (5.21,6.20) |
REF |
0.49 (-0.15,1.15) |
0.131 |
|
never >=3 cons n |
27 |
6.21 (5.38,7.15) |
0.031 |
31 |
6.16 (5.39,7.03) |
0.321 |
-0.05 (-1.27,1.15) |
0.938 |
|
<5 yr >= 3 cons n |
58 |
4.80 (4.36,5.29) |
0.228 |
51 |
5.61 (5.05,6.22) |
0.848 |
0.80 (0.07,1.56) |
0.033 |
|
>= 5yr >= 3 cons n |
179 |
4.98 (4.71,5.26) |
0.407 |
196 |
5.36 (5.08,5.65) |
0.260 |
0.38 (-0.02,0.78) |
0.064 |
aEstimated differences (with 95 % CI) were analyzed using linear regression. Adjustments were made for age at saliva test and familiar breast cancer. P-values ≤ 0.05 are indicated in bold.
Table 3: Effects of shift work and cancer status on DNA methylation of CpG1 in the MTNR1A gene.
4. Discussion
Epigenetic mechanisms such as DNA methylation, micro/ncRNA and histone modifications are promising biomarkers of disease development, progression and prognosis for several cancer types including breast cancer [35, 36]. Among the epigenetic factors 5mC methylation at CpG dinucleotides in the regulatory (promoter and enhancer regions) and also gene bodies (exons and introns) is the most studied biomarker. The rationale is that hypo- or hypermethylation of CpG sequences in the promoter regions alter gene transcription; increasing, decreasing or even causing gene silencing [37]. In this study, we therefore analyzed the methylation levels of 5mC in targeted CpG sequences in five receptor genes that are biologically relevant for investigation of the etiology of female breast cancer in general, and in shift work-related breast cancer in particular. The overall methylation levels varied in the five genes and as expected [38, 39] inter-individual variations across the genes were similar in all subjects regardless of case-control status. The lowest average MI was observed for ESR1 and the highest for MTNR1B. This variation in methylation levels may be due to biological factors rather than the number of CpGs since the number of CpGs for ESR1 (n=7) and MTNR1B (n= 6) genes were similar. A significantly higher methylation level was observed in the target promoter region of the MTNR1A gene in breast cancer cases compared to controls. No differences in methylation levels were observed between cases and controls for the other analyzed receptor genes. It has previously been shown that changes in DNA methylation patterns may be associated with changes in MTNR1A expression [40]. Since the anti-cancer actions of the circadian melatonin signal in human breast cancer cell lines and xenografts heavily involve MTNR1A-mediated mechanisms, changes in MTNR1A expression could play a role in the development of breast cancer [41]. Furthermore, melatonin suppresses ESR1 expression and ESR1 transcriptional activity via the MTNR1A receptor in ESR1-positive human breast cancer. Melatonin also regulates transactivation of estrogen-metabolizing enzymes, expression of core circadian clock and clock-related genes [41].
Analysis of individual CpGs in the MTNR1A gene further showed that increased methylation in CpG 1 was explanatory for the observed increased in breast cancer associated with higher methylation levels in the MTNR1A gene. Bioinformatic analyses showed that several central transcription factors such as CCCTC-binding factor (CTCF), GC-rich sequence DNA-binding factor (GCF), enhancer of zeste homolog 2 (EZH2), RAD21 cohesion complex component (RAD21), and tumor protein p53 (TP53) may bind to the MNTR1A promoter sequences. All of these transcription factors are implicated in breast carcinogenesis through different biological pathways such as DNA repair (RAD21and TP53) and epigenetic regulation of transcription (CTCF, GCF and EZH2). Long-term shift work with frequently repeated exposure to LAN may alter DNA methylation patterns, and genome-wide DNA methylation analysis has shown multiple alterations, where 3593 CpG sites were hypermethylated and 1816 CpG sites hypomethylated in long-term shift workers [42]. Disruption of the circadian nocturnal melatonin signal due to exposure to LAN promotes growth, metabolism, and signaling of human breast cancer which drives breast tumors to endocrine and chemotherapeutic resistance [41].
In this study, cases had higher MTNR1A CPG1 methylation levels than controls in the group of workers with intermediate exposure to night work, however, this effect was not observed for subjects in the high exposure group. Although our findings identifies MTNR1A methylation as a risk factor for breast cancer, the dependency of shift work cannot be conclusively confirmed. Our study is limited in that the methylation levels are generally low and that the number of subjects in each shift work group are low, thus, the data need to be confirmed in larger epidemiological studies. The present study is novel in the sense that it uses relatively well-matched groups of breast cancer cases and controls with night work exposure to investigate 5mC methylation levels at specific and biologically functional CpG sites. Pyrosequencing is the gold standard for DNA methylation analysis, permitting analysis of the target promoter region at each of the candidate genes. Additionally, pyrosequencing allows quantitative analysis of 5mC levels at each individual CpG dinucleotide. In contrast to global and random genome methylation methods, targeted and site specific approach enable selection of putative biologically functional target sites using the bioinformatics tools for binding motifs of transcription factors [43, 44]. Altogether, this study suggests that epigenetic regulation of MTNRIA may contribute to increased breast cancer, and functional studies are warranted to investigate the correlation between 5mC levels and MTNR1A expression.
Acknowledgements
We thank Mrs. Kristine Haugen Anmarkrud for excellent technical assistance in isolation and organization of DNA samples. We are also grateful to the nurses participating in the project.
Declaration of Interest
Ethics approval and consent to participate. Both cases and controls gave full informed written consent that their information could be used and published for research purposes, given that their personal details would remain anonymous. The Data Inspectorate and the Ethical Committee for Medical Research (REK-Sør) have evaluated and approved (project No. 6.2008/918) to conduct the study and the establishment of a biobank (registration No. 2361, FHI).
Consent for Publication
Not applicable.
Availability of Data and Materials
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing Interests
The authors declare that they have no conflict of interests.
Funding
This project was supported by the Norway Grants, under the Polish-Norwegian Research Program (Grant no. Pol-Nor/196940/22/2013-clockshift). JSE acknowledges a postdoctoral fellowship from NIOH.
Authors’ Contributions
JSE was involved in execution of the study, interpreted the results and drafted the initial version of the manuscript. ØS performed the statistical analysis. MPØ was involved in pyrosequencing, analysis and interpretation of the laboratory data. HØN analyzed CpG data and performed pyrosequencing. JAL established the case-control study and participated in the recruitment of subjects, the collection of data and establishment of the biobank, evaluated exposure metrics and contributed in writing the manuscript. KK participated in the establishment of case-control study, recruitment of subjects and evaluation of the exposure metrics. ER and BP were involved in the study design and helped in drafting the manuscript. SZ planned the study and was a major contributor in drafting and finalizing the manuscript. All authors read and approved the final manuscript.
References
- Martin AM, Weber BL. Genetic and hormonal risk factors in breast cancer. Journal of the National Cancer Institute (2000): 1126-1135.
- Lie JA, Andersen A, Kjaerheim K. Cancer risk among 43000 Norwegian nurses. Scandinavian Journal of Work, Environment & Health (2007): 66-73.
- Peng S, Lu B, Ruan W, et al. Genetic polymorphisms and breast cancer risk: evidence from meta-analyses, pooled analyses, and genome-wide association studies. Breast Cancer Research and Treatment (2011): 309-324.
- Zhang B, Beeghly-Fadiel A, Long J, et al. Genetic variants associated with breast-cancer risk: comprehensive research synopsis, meta-analysis, and epidemiological evidence. Lancet Oncology (2011): 477-488.
- Poli A, Marangoni F, Visioli F. Alcohol consumption and breast cancer risk. JAMA (2012):666.
- Pukkala E, Martinsen JI, Lynge E, et al. Occupation and cancer - follow-up of 15 million people in five Nordic countries. Acta Oncology (2009): 646-790.
- Weiderpass E, Meo M, Vainio H. Risk factors for breast cancer, including occupational exposures. Safety and Health at Work (2011): 1-8.
- Megdal SP, Kroenke CH, Laden F, et al. Night work and breast cancer risk: a systematic review and meta-analysis. European Journal of Cancer (2005): 2023-2032.
- Lie JA, Kjuus H, Zienolddiny S, et al. Night work and breast cancer risk among Norwegian nurses: assessment by different exposure metrics. American Journal of Epidemiology (2011): 1272-1279.
- Hansen J, Lassen CF. Nested case-control study of night shift work and breast cancer risk among women in the Danish military. Occupational and Environmental Medicine (2012): 551-556.
- Hansen J, Stevens RG. Case-control study of shift-work and breast cancer risk in Danish nurses: impact of shift systems. European Journal of Cancer (2012): 1722-1729.
- Pukkala E, Helminen M, Haldorsen T, et al. Cancer incidence among Nordic airline cabin crew. International Journal of Cancer (2012): 2886-2897.
- Wegrzyn LR, Tamimi RM, Rosner BA, et al. Rotating night-shift work and the risk of breast cancer in the nurses' health studies. American Journal of Epidemiology (2017): 532-540.
- Cordina-Duverger E, Menegaux F, Popa A, et al. Night shift work and breast cancer: a pooled analysis of population-based case–control studies with complete work history. European Journal of Epidemiology (2018): 369-379.
- Liu W, Zhou Z, Dong D, et al. Sex differences in the association between night shift work and the risk of cancers: a meta-analysis of 57 articles. Disease Markers (2018): 7925219.
- Vaughn CB, Freudenheim JL, Nie J, et al. Sleep and breast cancer in the Western New York Exposures and Breast Cancer (WEB) study. Journal of Clinical Sleep Medicine (2018): 81- 86.
- Bustamante-Montes LP, Flores-Meza B, Hernández-Valero MA, et al. Night shift work and risk of breast cancer in women. Archives of Medical Research (2019): 393-399.
- Szkiela M, Kusidel E, Makowiec-Dabrowska T, et al. Night shift work-a risk factor for breast cancer. International Journal of Environmental Research and Public Health (2020).
- Ward EM, Germolec D, Kogevinas M, et al. Carcinogenicity of night shift work. Lancet Oncology (2019): 1058- 1059.
- Travis RC, Balkwill A, Fensom GK, et al. Night shift work and breast cancer incidence: Three prospective studies and meta-analysis of published studies. Journal of the National Cancer Institute (2016).
- Son H, Kang Y. Breast cancer screening among shift workers: a nationwide population-based survey in Korea. International Journal of Occupational and Environmental Health (2017): 94-97.
- Jones ME, Schoemaker MJ, McFadden EC, et al. Night shift work and risk of breast cancer in women: the Generations Study cohort. British Journal of Cancer (2019): 172-179.
- Pham T-T, Hwang M, Lee E-S, et al. Night-shift work and risk of breast cancer in Korean women. Clinical Epidemiology (2019): 743-751.
- Stevens RG. Testing the light-at-night (LAN) theory for breast cancer causation. Chronobiology International (2011): 653-656.
- Haus EL, Smolensky MH. Shift work and cancer risk: potential mechanistic roles of circadian disruption, light at night, and sleep deprivation. Sleep Medicine Reviews (2013): 273-284.
- Hardeland R, Madrid JA, Tan DX, et al. Melatonin, the circadian multioscillator system and health: the need for detailed analyses of peripheral melatonin signaling. Journal of Pineal Research (2012): 139-166.
- Sanchez-Hidalgo M, Guerrero JM, Villegas I, et al. Melatonin, a natural programmed cell death inducer in cancer. Current Medicinal Chemistry (2012): 3805-3821.
- Langley AR, Graham CH, Grundy AL, et al. A cross-sectional study of breast cancer biomarkers among shift working nurses. BMJ Open (2012): e000532.
- Roy D, Belsham DD. Melatonin receptor activation regulates GnRH gene expression and secretion in GT1-7 GnRH neurons. Signal transduction mechanisms. Journal of Biological Chemistry (2002): 251-258.
- Alvarez-Garcia V, Gonzalez A, Martinez-Campa C, et al. Melatonin modulates aromatase activity and expression in endothelial cells. Oncology Reports (2013): 2058-2064.
- Bai Z, Gust R. Breast cancer, estrogen receptor and ligands. Archiv der Pharmazie (Weinheim) (2009):133-149.
- Hill SM, Huettner F, Murray J, et al. Contribution of breast density to the volume of the augmented breast: A preliminary study. Canadian Journal of Plastic Surgery (2011): 93-96.
- Carugno M, Maggioni C, Crespi E, et al. Night shift work, DNA methylation and telomere length: An investigation on hospital female nurses. International Journal of Environmental Research and Public Health (2019): 2292.
- Messeguer X, Escudero R, Farre D, et al. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics (2002): 333-334.
- Davalos V, Martinez-Cardus A, Esteller M. The epigenomic revolution in breast cancer: from single- gene to genome-wide next-generation approaches. American Journal of Pathology (2017): 2163-2174.
- Flavahan WA, Gaskell E, Bernstein BE. Epigenetic plasticity and the hallmarks of cancer. Science (2017).
- Esteller M. Epigenetics in cancer. New England Journal of Medicine (2008): 1148-1159.
- Guntrum M, Vlasova E, Davis TL. Asymmetric DNA methylation of CpG dyads is a feature of secondary DMRs associated with the Dlk1/Gtl2 imprinting cluster in mouse. Epigenetics Chromatin (2017): 31.
- Tejedor JR, Fraga MF. Interindividual epigenetic variability: Sound or noise? Bioessays (2017).
- Kim B, Rincon Castro LM, Jawed S, et al. Clinically relevant concentrations of valproic acid modulate melatonin MT(1) receptor, HDAC and MeCP2 mRNA expression in C6 glioma cells. European Journal of Pharmacology (2008): 45-48.
- Hill SM, Belancio VP, Dauchy RT, et al. Melatonin: an inhibitor of breast cancer. Endocrine-Related Cancer (2015): R183-R204.
- Zhu Y, Stevens RG, Hoffman AE, et al. Epigenetic impact of long-term shiftwork: pilot evidence from circadian genes and whole-genome methylation analysis. Chronobiology International (2011): 852-861.
- Weinstock GM. ENCODE: more genomic empowerment. Genome Research (2007): 667-668.
- Harrington CT, Lin EI, Olson MT, et al. Fundamentals of pyrosequencing. Archives of Pathology & Laboratory Medicine (2013): 1296-1303.


 Impact Factor: * 4.1
Impact Factor: * 4.1 Acceptance Rate: 74.74%
Acceptance Rate: 74.74%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 