Obstruction Syndrome and Septic Shock Induced By etoposide explained by Pharmacogenetics a Case Report
CHERIFI François1,2*, SASSIER Marion3, BOURGINE Joanna3, DA SILVA Angelique1, JOLY Florence2
1Breast Cancer Unit, François Baclesse Center, Normand du Sein Institute, Caen, France
2Clinical Research Department, Centre François Baclesse, Caen, France
3Caen University Hospital Center, CHU Caen, Pharmacology, France
*Corresponding Author: CHERIFI François, Caen University Hospital Center | CHU Caen Pharmacology, Doctor of Pharmacy, France
Received: 12 March 2022; Accepted: 29 March 2022; Published: 19 April 2022
Article Information
Citation:
François CHERIFI, SASSIER Marion1, Bourgine Joanna, DA SILVA Angelique, Joly Florence. Obstruction Syndrome and Septic Shock Induced By etoposide explained by Pharmacogenetics A Case Report. Journal of Cancer Science and Clinical Therapeutics 6 (2022): 207-213.
View / Download Pdf Share at FacebookKeywords
<p>Prognosis; Leucopenia; Toxicities</p>
Article Details
1. Introduction
Seminoma are common in young men. Seminoma have good prognosis with 10-year survival rate of more than 95% [1, 2]. After surgery, the standard treatment of seminoma is the chemotherapeutic combination of bleomycin, etoposide and cisplatin (BEP). The most usual and severe adverse events (AEs) observed with BEP are leucopenia (9% grade 3-4), febrile neutropenia (15%), neurotoxicity (6% grade 2-3) and renal (4% grade 1-3) [3]. Etoposide limiting toxicity is most frequently leucopenia but rare digestive toxicities exist. These toxicities are mainly dose-dependent and can occur in case of drug overdose due to drug-drug interactions (DDI) [4, 5]. Frequency and severity of AEs are related to the capacity of each individual to metabolize or transport drugs and may differ significantly according to genetic factors [6]. Here we report an unusual case of ileal obstruction syndrome complicated with a septic shock due to pharmacogenetics, in a patient treated with cisplatin and etoposide for a seminoma.
2. Case Report
A 53-year-old man was diagnosed with seminoma stage III-B with large lumbar-aortic ganglionic extension. His past medical history included dyslipidemia, diabetes, hypertension, obesity and sleep apnea. His usual treatment included irbesartan, hydrochlorothiazide, spironolactone, nicardipine, rilmenidine, atorvastatin, aspirin and esomeprazole. After orchidectomy, he received etoposide (100mg/m² day 1 to 5) and cisplatin (20mg/m² day 1 to 5) (EP) every 3 weeks. Bleomycin was not administered considering the comorbidities. One week after the first course of EP, the patient was hospitalized for an ileal obstruction syndrome associated with grade 4 neutropenia in CTCAE score (Common Terminology Criteria for Adverse Events). It was complicated by septic shock and an acute renal failure. After laparotomy, the patient was transferred to an Intensive Care Unit (ICU). The evolution was progressively favorable after intubation, vascular filling associated with amine support and antibiotic therapy (piperacilline-tazobactam, amikacine). The patient was discharged of ICU after twelve days. Due to unexpected AEs we suspected DDI induced by etoposide, a substrate of hepatic cytochromes P-450 isoforms 3A4 and 3A5 (CYP3A4 and CYP3A5) and P-glycoprotein (P-gp) and can interact with CYP3A4 and P-gp inhibitors. The usual treatment of our patient contained several inhibitors of P-gp (atorvastatin, nicardipine and irbesartan) which could have increased concentration of etoposide, explaining the severe AEs observed, although etoposide levels were not tested in our case. Therefore, the treatment was optimized before the administration of the second course. Atorvastatin, nicardipine and irbesartan were switched respectively to pravastatin, amlodipine and perindopril. Then the same chemotherapy regimen was rechallenged with identical doses and the patient presented after the 2nd and the 3rd courses a major digestive toxicity with grade 3 emesis, abdominal pain and grade 4 neutropenia complicated by neutropenic colitis requiring hospitalization. The scanner showed intestinal inflammation without organic obstruction. We achieved asymptomatic treatment with analgesic, antiemetic and antibiotic therapy. The patient was discharged 6 and 28 days later, respectively, without any surgery. Due to these severe toxicities, chemotherapy was stopped after the third course while the patient still remained in complete response.
3. Management and Outcome
The registration number to the French pharmacovigilance center was CN20180967. The assessment of causality by the Naranjo scale found a score of 11 (a score ≥ 9 signs a definite reaction), the value of causality assessment according to the French method was probable and confirms the implication of EP in the occurrence of the AE observed [7, 8] (supplementary data 1). To investigate these severe unusual and repeated AEs occurring at standard label doses, the polymorphisms of genes involved in metabolisms and drug transport of etoposide were analyzed. Therefore CYP3A5, CYP3A4, ABCB1, POR and UGT1A1 single-nucleotide polymorphisms (SNPs) were assessed using real-time polymerase chain reaction (PCR) Taqman assays and DNA sequencing technologies. The analysis revealed the presence of several polymorphisms in the studied CYP3A4, CYP3A5, POR and ABCB1 genes: CYP3A4*1/*22 (15389C>T; rs35599367), CYP3A5*3/*3 (6986A>G; rs776746), POR*1/*28 (1508C>T; rs1057868) and ABCB1 (1236T/T, rs1128503; 2677T/T, rs2032582; 3435T/T, rs1045642). None of the polymorphisms searched on UGT1A1 had been identified (Table 1). This genetic profile could explain the drug accumulation especially for etoposide, and thus the observed severity of AEs.
|
Gene |
Polymorphism |
dbSNP ID |
Patient |
|
CYP3A4 |
-392A>G (*1B) |
rs2740574 |
AA |
|
15389 C>T (*22) |
rs35599367 |
CT |
|
|
CYP3A5 |
6986 A>G (*3) |
rs776746 |
GG |
|
12952 C>T (*6) |
rs10264272 |
CC |
|
|
27131 insT (*7) |
rs41303343 |
Absence |
|
|
ABCB1 |
1236 C>T |
rs1128503 |
TT |
|
2677 G>T/A |
rs2032582 |
TT |
|
|
3435 C>T |
rs1045642 |
TT |
|
|
POR |
1508 C>T (*28) |
rs1057868 |
TT |
|
UGT1A1 |
TA repeat (*28) |
rs8175347 |
(TA)6/(TA)6 |
|
c.211G>A (*6) |
rs4148323 |
GG |
|
|
c.-3156G>A (*93) |
rs10929302 |
GG |
|
|
c.-3263T>G (*60) |
rs4124874 |
TT |
|
|
c.686C>A (*27) |
rs35350960 |
CC |
Table 1: Polymorphisms in phase I and phase II metabolic enzymes and in drug transporter genes affecting pharmacokinetics of etoposide.
We can see the different polymorphisms searched in the database, in bold the identified polymorphism. We didn’t find polymorphisms on UGT1A1. dbSNP ID: Single Nucleotide Polymorphism Database Discussion: Serious AEs induced by etoposide are uncommon and conventionally seen when used in high doses or with DDI. Other circumstances are rare and polymorphisms-caused AEs are very poorly described in the literature and need more exploration [9]. AEs developed by our patient (mainly digestive and hematological) were in line with those associated with etoposide, and did not match with usual cisplatin AEs. Therefore, we did not perform the search of Glutathione-S-transferases (GSTs) polymorphisms, involved in the metabolic pathway of cisplatin. Nevertheless, cisplatin could have worsened the toxicity. Etoposide is a commonly used semisynthetic derivative of podophyllotoxin that exhibits antitumor activity by stabilizing the double-stranded DNA cleavage normally catalyzed by topoisomerase II and thus by inhibiting faithful religation of DNA breaks. These double-strand DNA breaks subsequently trigger the desired antitumor effects of the drug [10]. Etoposide is mainly metabolized by hepatic CYP3A4 and CPY3A5 into a catechol metabolite. Etoposide also undergoes glutathione and glucuronide conjugation which are catalyzed by GSTT1/GSTP1 and UGT1A1, respectively
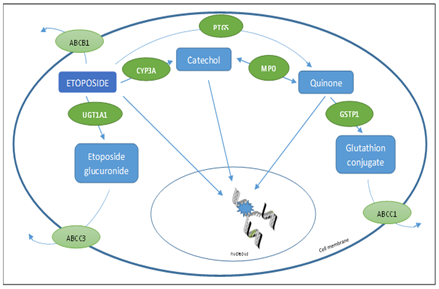
Figure 1: Enzymes involved in the metabolic pathway of etoposide, adapted from de Wit, R and al [3].
Etoposide is mainly metabolized by hepatic CYP3A4 and CPY3A5 into a catechol metabolite. Etoposide undergoes glutathione and glucuronide conjugation which are catalyzed by GSTT1/GSTP1 and UGT1A1. The ABC protein, an efflux transporter family, will try to clear out etoposide of the cell In our case we found several polymorphisms of metabolism and transport pathways of etoposide. First our patient had a CYP3A4 heterozygous deficient variant *22 and a CYP3A5 homozygous deficient variant *3. The CYP3A subfamily enzymes play a major role in the metabolism of clinically used drugs. Only a few SNPs are known to influence CYP3A4 enzyme function. Only one intronic variant (rs35599367) allocated to *22 was shown to be associated with decreased CYP3A4 activity[11]. CYP3A4*22 leads to 50% reduction of mRNA expression and, subsequently to a reduced enzyme activity. The relevance of CYP3A4*22 is even more important in subjects having no active CYP3A5 enzyme [11], like in our case. Indeed CYP3A5*3 leads to aberrant splicing and a truncated protein and result in non-functional proteins [12]. It has a frequency ranging in up to 88-97% in Caucasian populations [13]. In the gene polymorphism analysis we also found an homozygous polymorphism *28 of cytochrome P450 oxidoreductase (POR) which could be associated with an increase in metabolic activity of CYP450 [14]. However, in the absence of expression of CYP3A5 (CYP3A5*3/*3 genotype), its effect becomes negligible. Finally, the analysis revealed a polymorphism of ABCB1 (ATP binding cassette subfamily B member 1). The human multidrug-resistance MDR1 gene, an efflux transporter also known as ABCB1, encodes for the efflux transporter P-glycoprotein (P-gp). This protein acts as a primary active transporter, playing an important role in protecting tissues from toxic xenobiotics and endogenous metabolites. The degree of expression and function of ABCB1 could directly influence the therapeutic effectiveness of such agents, affecting their bioavailability [15]. The ABCB1 gene has been extensively studied for its characteristic polymorphisms, with at least 50 SNPs identified. Three polymorphisms frequently studied for their effects on P-gp expression, functionality, and substrate distribution are 1236C>T (rs1128503), 2677G>T/A (rs2032582), and 3435C>T (rs1045642). In Caucasians, the frequency of the 1236T/T, 2677T/T and 3435T/T genotypes is respectively 38%, 19% and 28% [16]. These three polymorphisms were present in our patient. Hoffmeyer et al. reported that the 3435T/T genotype was correlated with reduced expression of P-gp and, therefore, reduced cellular elimination of chemotherapeutic drugs leading to higher plasma concentrations [17]. Furthermore hematopoietic stem cells and intestinal epithelial cells are reported to express P-gp, thus higher drug concentrations in homozygous 3435T/T genotype could lead to more frequent grade 3–4 neutropenia, febrile neutropenia, and diarrhea [18], especially when associated with drug-drug interaction which worsen the metabolic situation. This can explain a slightly better tolerance of the two last cures of chemotherapy after optimization of the patient’s usual treatment.
Lavanderos et al. also analyzed, in a prospective study, the genetic polymorphisms of BEP-treated testicular cancer patients. They showed that specific ERCC1/2, MDR1, GSTP1, and BLMH variant were associated with severe leukopenia, vomiting, nausea and alopecia [19].
This study reinforces the impact between genetic variants and AEs. However, their findings are more relevant for cisplatin and bleomycin related toxicity and did not impact directly etoposide. Besides, they did not describe an increased risk of digestive toxicities like in our case. Our patient presented several genetic variants impairing etoposide clearance, worsened by DDI which caused an overdose responsible of serious adverse effects. Unfortunately, pharmacokinetic assays had not been performed to confirm our hypothesis. Clinicians have to be aware of adverse effects originating from polymorphisms and the recommendations from the French Network of Pharmacogenetics (RNPGx) who encourages the use of pharmacogenetics evaluation to offer the possibility of predictive personalized medicine and its corollary of reduced cost and lower risk of iatrogenic adverse effects [20], especially when unusual AEs occur in the field of oncology. In such cases potential DDI should be carefully reviewed and a close monitoring of plasma levels of chemotherapy has to be performed. This finding is particularly relevant because it allows clinicians to adapt chemotherapy dosage or switch to another protocol.
3. Conclusion
Genetic polymorphisms are a relevant factor in cancer drug pharmacokinetics, affecting metabolism, drug exposure and could explain severe AEs occurring at standard label doses. Pharmacogenetics analysis can give a comprehensive insight in case of unexplained and severe AEs and should be performed.
Consent to Participate
Informed and written consent was obtained from the patient for the publication of his case.
Conflicts of Interest
We have no Conflict of Interest to declare
References
- Williams S, Birch R, Einhorn L, et al. Treatment of disseminated germ-cell tumors with cisplatin, bleomycin, and either vinblastine or etoposide. N Engl J Med 316 (1987): 1435-1440.
- Oing C, Seidel C, von Amsberg G, et al. Pharmacotherapeutic treatment of germ cell tumors: standard of care and recent developments. Expert Opin Pharmacother 17 (2016): 545-560.
- De Wit R, Roberts T, Wilkinson PM. Equivalence of 3 or 4 cycles of BEP chemotherapy administered on a 3 or 5 day schedule in good prognosis germ cell cancer, a randomized study of the European Organisation for Research and Treatment of Cancer Genitourinary Tract Cancer Cooperative Group an. J Clin Oncol 19 (2001): 2001-1640.
- McLeod HL. Clinically relevant drug–drug interactions in oncology. British Journal of Clinical Pharmacology 45 (1998): 539-544.
- Canal P, Chatelut E, Guichard S. Practical Treatment Guide for Dose Individualisation in Cancer Chemotherapy: Drugs 56 (1998): 1019-1038.
- Phan VH, Moore MM, McLachlan AJ, Piquette-Miller M, Xu H, Clarke SJ. Ethnic differences in drug metabolism and toxicity from chemotherapy. Expert opinion on drug metabolism & toxicology 5 (2009): 243-257.
- Naranjo CA, Busto U, Sellers EM, et al. A method for estimating the probability of adverse drug reactions. Clin Pharmacol Ther 30 (1981): 239-245.
- Moore N, Berdaï D, Blin P, et al. Pharmacovigilance – The next chapter. Therapies 74 (2019): 557-567.
- Del Re M, Latiano T, Fidilio L, et al. Unusual gastrointestinal and cutaneous toxicities by bleomycin, etoposide, and cisplatin: a case report with pharmacogenetic analysis to personalize treatment. EPMA Journal 8 (2017): 69-73.
- Yang J, Bogni A, Schuetz E. Etoposide pathway. Pharmacogenet Genomics 19 (2009): 552-553.
- Werk A, Cascorbi I. Functional gene variants of CYP3A4. Clin Pharmacol Ther 96 (2014): 340-348.
- Wang D, Guo Y, Wrighton A, et al. Intronic polymorphism in CYP3A4 affects hepatic expression and response to statin drugs. The pharmacogenomics journal 11 (2011): 274-286.
- Zanger U, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther 138 (2013): 103-141.
- Zhang H, Li Z, Liu J, et al. Correlation of Cytochrome P450 Oxidoreductase Expression with the Expression of 10 Isoforms of Cytochrome P450 in Human Liver. Drug Metab Dispos 44 (2016): 1193-200.
- Diekstra MHM, Swen JJ, Boven E, et al. CYP3A5 and ABCB1 polymorphisms as predictors for sunitinib outcome in metastatic renal cell carcinoma. Eur Urol 68 (2015): 621-629.
- Hamidovic A, Hahn K, Kolesar J. Clinical significance of ABCB1 genotyping in oncology. J Oncol Pharm Pract 16 (2010): 39-44.
- Hoffmeyer S, Burk O, Richter O von, et al. Functional polymorphisms of the human multidrug-resistance gene: multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proceedings of the National Academy of Sciences 97 (2000): 3473-3478.
- Chaudhary P, Roninson IB. Expression and activity of P-glycoprotein, a multidrug efflux pump, in human hematopoietic stem cells. Cell 66 (1991): 85-94.
- Lavanderos MA, Cayún JP, Roco Á, et al. Association study among candidate genetic polymorphisms and chemotherapy-related severe toxicity in testicular cancer patients. Frontiers in Pharmacology 10 (2019): 1-10.
- Picard N, Boyer J-C, Etienne-Grimaldi M-C, et al. Pharmacogenetics-based personalized therapy: Levels of evidence and recommendations from the French Network of Pharmacogenetics (RNPGx). Therapies 72 (2017): 185-192.


 Impact Factor: * 4.1
Impact Factor: * 4.1 Acceptance Rate: 74.74%
Acceptance Rate: 74.74%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 