Obtaining a Recombinant Protein Based on the E7 Antigen of Human Papillomavirus Type 18
Rodriguez Rodriguez N1*, Alfonso Chaviano AB1, Granadillo Rodríguez M1, Batte Figueredo A1, Torrens I1
1Department of Cancer, Center for Genetic Engineering and Biotechnology, P.O. Box 6162, Cubanacan, Playa, Havana 10600, Cuba
*Corresponding Author: Rodriguez Rodriguez, Department of Cancer, Center for Genetic Engineering and Biotechnology, P.O. Box 6162, Cubanacan, Playa, Havana 10600, Cuba
Received: 15 December 2021; Accepted: 21 February 2022; Published: 26 August 2022
Article Information
Citation: Rodriguez Rodriguez N, Alfonso Chaviano AB, Granadillo Rodríguez M, Batte Figueredo A, Torrens I. Obtaining a Recombinant Protein Based on the E7 Antigen of Human Papillomavirus Type 18. Journal of Cancer Science and Clinical Therapeutics 6 (2022): 336-343.
View / Download Pdf Share at FacebookAbstract
Background: Cervical cancer is the fourth most common cancer in women worldwide and was the leading cause of death among approximately 300 000 cases in 2018. The main risk factor for developing this disease is the persistent infection of high-risk types of Human Papillomavirus (HPV); specifically, HPV-18 is recognized as one of the major contributors for adenocarcinoma and squamous cancer. There are three prophylactic vaccines to prevent infection by HPV, but these vaccines are not effective in infected people. On the other hand, despite the success of various types of therapeutic vaccine candidates in clinical trials, none of them is commercially available to treat women with HPV-related malignancies. As the methods used for obtaining those therapeutic candidates are awfully expensive, they could be inaccessible for developing countries. In this scenario, E7 antigen of HPV is considered an ideal target for developing therapeutic vaccines. In accordance with this, the aim of this work is to obtain a recombinant fusion protein with high levels of purity through a profitable process, which could be used as therapeutic alternative for treating tumors expressing the E7 antigen of HPV-18.
Results: We have obtained the novel recombinant protein CIGB550-E718 that comprises the fusion between an E7 mutein of HPV-18 and the cellpenetrating peptide with immunostimulatory properties CIGB550. The expression of the fusion protein evaluated using two strains of E. coli and 16 culture media with different composition enabled us to choose the best setting in which the interest protein accounted for approximately 16% of the total cellular protein. The best results were obtained using BL21(DE3) cells as host system grown in a culture medium containing M9 salt, casein hydrolysate and glycerol as a carbon source. The recombinant protein was purified by a single affinity chromatographic
Keywords
<p>Human papillomavirus; Recombinant protein; Mutein; Oncoprotein</p>
Article Details
Abbreviations:
HPV: Human papillomavirus; CTL: Cytotoxic T lymphocyte; CPP: Cell-penetrating peptides; MHC-I: Major histocompatibility complex class I; PCR: Polymerase chain reaction; pRb: Retinoblastoma protein; SDS-PAGE: Sodium dodecyl sulphate–polyacrylamide gel electrophoresis; MW: Molecular weight
1. Background
Cervical cancer is the fourth most common cancer in women worldwide and was the leading cause of death among approximately 300 000 cases in 2018 [1]. The development of this disease is usually associated with the persistent infection by high-risk viral types of Human Papillomavirus (HPV). Specifically, HPV-18 together with HPV-16 causes up to 70% of invasive cervical cancer in global female population [2] and it is recognized as one of the major contributors for adenocarcinoma and squamous cancer [3]. There are three prophylactic vaccines to prevent HPV infection; however, these vaccines do not have therapeutic effects in infected women. Several therapeutic vaccine candidates are currently undergoing clinical trials and thus, are yet to reach the market [4]; and even if shown to be effective, there are no guarantees that the required manufacturing processes are cheap enough to make them accessible to patients in developing countries, where the highest incidences are found [5,6]. HPV E6 and E7 viral oncoproteins play the pivotal role in driving the cells toward oncogenesis and together are co-expressed in most of cervical cancer [7]. Accordingly, both proteins are considered as ideal antigenic targets for developing therapeutic vaccines and immunotherapeutic strategies against tumors associated with HPV infection since they are consistently expressed in HPV-associated malignancies and precancerous lesions, playing crucial roles in the generation and maintenance of HPV-associated disease [8,9]. Among all the HPV oncogenes, E7 was the first oncogene to be discovered and, compared to E6, it is the most widely used due to its higher expression, it is better conserved among the different types of HPV and it is well characterized immunologically in preclinical models [10]. Unfortunately, exogenous proteins are weak immunogens that normally induce a humoral immune response and have limited efficacy to generate a potent Cytotoxic T Lymphocyte (CTL) response which is necessary for the success of the vaccination. To overcome this limitation, Cell-Penetrating Peptides (CPP) could be used [11]. The vaccine candidate CIGB550-E7 was designed by Granadillo and co-workers for treating women with lesions caused by HPV-16 [12]. This candidate comprises the fusion between a cell-penetrating and immunostimulatory peptide (CIGB550) and the E7 antigen of HPV-16. The preclinical results obtained with this candidate demonstrate the ability of the above mentioned CPP to enter the E7 antigen into antigen-presenting cells favoring the antigen presentation in the context of the Major Histocompatibility Complex Class I (MHC-I) [12]. Considering the need for a therapeutic vaccine against tumors expressing the E7 antigen of HPV-18 and making use of CPP as a technological platform, this paper describes the design, expression, and purification of the novel recombinant protein CIGB550-E718. We designed a mutant antigen based on the E7 protein of HPV-18 fused to the CPP CIGB550 that was expressed in two strains of E. coli using 16 culture media with different composition, and was further purified by a single chromatographic affinity step. Yields of this non-optimized process support a promising scenario for the future production and scale-up of the CIGB550-E718.
2. Results
2.1. Construction of the plasmid pABA
The sequence of the gene expressing the E7 mutein assembled by Polymerase Chain Reaction (PCR) is shown in the Figure 1A. The recognition sites for HindIII and BamHI restriction enzymes are included at the beginning and at the end of this sequence, respectively. In addition, two replacements by glycine were done, both contained in the L25X26C27X28E29 domain. Mutations in this domain avoid E7 protein interaction with the retinoblastoma protein (pRb), being a regulatory advantage for the future development of a therapeutic vaccine based on the E7 oncoprotein [13, 14]. The band corresponding to the designed E7-18m gene had the expected size of 327 bp, as shown in Figure 1B. After enzymatic digestion of the vector pPEPE7M-7 and following insertion of the E7-18m gene, the E7 mutein was fused to the CPP CIGB550 obtaining the CIGB550-E718 gene into the new vector denominated pABA (Figure 2). Thirteen clones were selected, with twelve clones being positive to restriction analysis. These clones were sequenced and three out of 12 clones had the sequence corresponding to the recombinant protein CIGB550-E718 (Figure 2). The clone number two was selected for following assays.
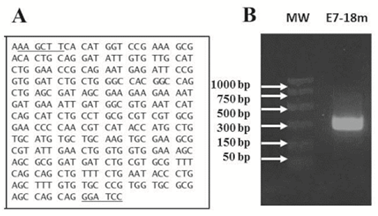
Figure 1: Synthesis of E7-18m gene expressing a mutein of HPV-18 E7 protein assembled by PCR. A. Sequence of the E7-18m gene. Underlined bases in the sequences correspond with the restriction sites for HindIII and BamHI enzymes. B. 1.2% agarose gel electrophoresis showing E7-18m gene.
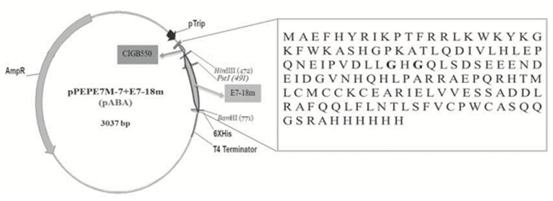
Figure 2: Schematic representation of pABA vector expressing the recombinant protein CIGB550-E718. Amino acids sequence of protein comprising a fusion between the CPP CIGB550 and a mutein of HPV-18 E7 protein followed by a 6xHis-tag is shown in the chart. Two bases replaced by glycine are represented in bold.
2.2. Expression of the fusion protein CIGB550-E718
coli BL21(DE3) and BL21-RP cells were transformed with the pABA vector and the expression of the recombinant protein was induced. The protein expression analysis by 15% sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) was made for BL21(DE3) and BL21-RP strains revealing a band at the expected molecular weight (MW) of approximately 20 kDa, which corresponds with the recombinant protein CIGB550-E718 (Figure 3A). The expression of the CIGB550-E718 accounted for approximately 14.7% of the total cellular protein in BL21(DE3) strain compared to only 4% in BL21-RP cells. Higher MW aggregates and a degradation of this protein were visualized by the Western Blot analysis (Figure 3B). Bl21(DE3) strain transformed with pABA plasmid was selected to evaluate the expression of the CIGB550-E718 in different culture media using two carbon sources: glucose and glycerol. As shown in Figure 4 panels A and B, the major band of 20 kDa was observed for all culture media evaluated using any of the carbon sources. In comparative terms, the best protein expression was achieved when glycerol was added to the culture media. The protein expression determined by densitometry was approximately 16% of total cellular protein in the M9 salt medium enriched with casein hydrolysate and glycerol (medium identified with the letter D). Statistical analysis (Figure 5) for each condition suggests that there are no significant differences among culture media containing glycerol (p>0.05). However, for culture media containing glucose, the expression of the recombinant protein was higher for the culture medium containing only M9 salt (identified with letter A) respect to the same medium enriched with tryptone, yeast extract and casein hydrolysate (p<0.05). Thus, taking in consideration these analyses, M9 salt medium enriched with casein hydrolysate and glycerol as a carbon source (medium identified with the letter D) was selected for protein purification process.
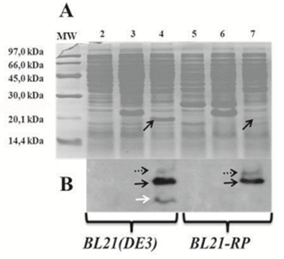
Figure 3: Expression of the recombinant protein CIGB550-E718. A. SDS-PAGE (15%). B. Western Blot. Lane 1: MW; lanes 2 and 5: E. coli BL21(DE3) or BL21-RP strains transformed with pM240 vector; lanes 3 and 6: E. coli BL21(DE3) or BL21-RP strains transformed with pPEPE7M-7 vector; lanes 4 and 7: E. coli BL21(DE3) or BL21-RP strains transformed with new pABA vector. Black arrows indicate the monomer of the CIGB550-E718. Dash black arrows correspond to protein aggregates, while white arrow shows a degradation of the same protein.
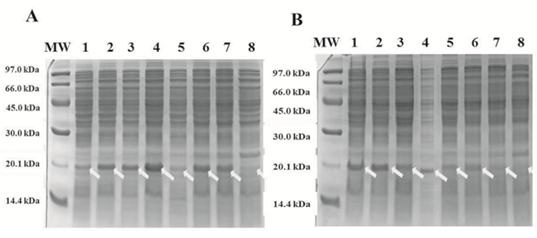
Figure 4: Expression of the protein CIGB550-E718 in 16 culture media using two carbon sources. SDS-PAGE (15%) A. Culture media with glycerol as carbon source. B. Culture media with glucose as carbon source. Lines 1-8: E. coli BL21(DE3) harboring pABA induced with 40 µg/mL β-indolacrylic acid in culture media from A to H (See Materials and Methods section). White arrows show the band corresponding to the CIGB550-E718.
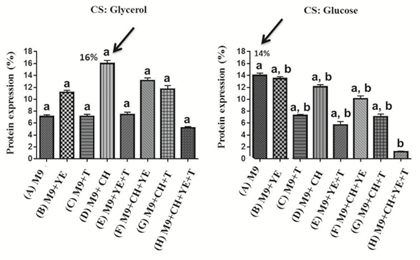
Figure 5: Percentage of the CIGB550-E718 expression in different culture media. Statistical analysis was induced to choose the best culture medium for expressing the recombinant protein. Three experimental replicates were analyzed. The average of the percentage of expression of each medium ± interquartile range is shown. Statistical differences were analyzed by Dunn test, p<0.05. Black arrow shows culture media corresponding to the best percentage of protein expression determined by densitometry. CS: carbon source, M9: M9 salt, YE: yeast extract, T: tryptone, CH: casein hydrolysate.
2.3. Purification of the recombinant protein CIGB550-E718
The non-optimized purification process of the CIGB550-E718 protein was carried out from 2.5 L of culture medium containing M9 salt, casein hydrolysate and glycerol (medium identified with letter D). About twelve grams of biomass per liter of induced culture were obtained at the end of the fermentation process. As shown in Figure 6A lane 2, the CIGB550-E718 accounted for about 16% of the total cellular protein, and migrates as an approximately 20 kDa protein in 15% SDS-PAGE. This molecule was found in the insoluble fraction after cell disruption (Figure 6A, lane 3). Therefore, a protein extraction step using 6 M urea was done in order to obtain the protein in the soluble fraction (Figure 6A, lane 6). The following phase consisted in a single chromatographic step of IMAC which allowed us to obtain the recombinant protein CIGB550-E718 with 90% purity (Figure 6A, lane 10). The immunoidentification analysis using an anti-HPV-18 E7 polyclonal antibody is shown in Figure 6B. A major band of approximately 20 kDa and aggregates of the CIGB550-E718 are revealed from eluates of IMAC and molecular exclusion (Figure 6B, lanes 9 and 10). Yields of the purification process were around 32 mg of the recombinant protein CIGB550-E718 per liter of induced culture.
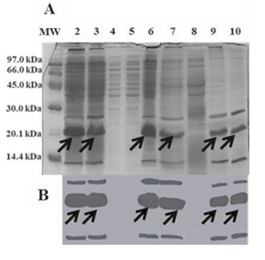
Figure 6: Expression and purification of the recombinant protein CIGB550-E718. A. SDS-PAGE (15%) B. Western blot. Lane 1: MW; lane 2: biomass containing the fusion protein accounting for approximately 16% of the total cellular protein; lanes 3 and 4: insoluble and soluble fractions after cell disruption, respectively; lanes 5 and 6: insoluble and soluble fractions after protein extraction, respectively. IMAC purification process; lane 7: initial sample; lane 8: wash with 10 mM imidazole; lane 9: elution with 300 Mm imidazole; lane 10: elution with 10 mM Tris pH 8 by molecular exclusion chromatography. Arrows show a monomer corresponding to the CIGB550-E718 protein.
3. Discussion
In this paper we describe the whole process, from design to purification of a novel recombinant protein (CIGB550-E718), which could be used to treat precancerous lesions and cancer in women infected with HPV-18. This molecule comprises the fusion between the CPP CIGB550, capable to enhance the immune response, and a mutein of the HPV-18 E7 protein. The mutant antigen design included the substitution of two amino acids (C27 and E29 by G), with the aim of suppressing the oncogenic potential of this molecule through its interaction with pRb [15, 16]. These substitutions were necessary due to existent prohibition for using non-modified oncogenes for human vaccination, and in order to reduce upcoming regulatory objections for the development of the future vaccine candidate CIGB550-E718. Valuable studies suggest that E7 antigen alone is not capable to penetrate into cells because the cellular membrane is impermeable to hydrophilic substances [17, 18]. In accordance to this evidence, we employed a CPP described by Granadillo et al. in 2011. This CPP has an intrinsic in vivo ability to enter into cells and it is also capable to internalize the exogenous E7 antigen [12, 19]. The covalent fusion of the mutein and the CPP CIGB550, should be essential for internalizing the E7 antigen of HPV-18 into antigen presenting cells for subsequent presentation in the MHC-I context. The antigenic presentation in this context may enhance the CTL response, necessary for the success of the vaccination with the CIGB550-E718. Furthermore, it is possible that the CPP is improving the E7 expression in E. coli, which allows us to obtain it with a high percentage respect to all proteins that the host is expressing.
The number of options when designing an expression system is considerably high. The steps flow to obtain the novel CIGB550-E718 was supported by a previous trial-and-error work when the CIGB550-E7 therapeutic vaccine candidate was obtained [20]. Choosing the perfect combination is not possible a priori, due to the possible combination variability between protein expression systems and growth conditions (medium, pH, Temperature, Induction). So, we decided to evaluate some conditions that allow us to obtain a high percentage of the recombinant protein CIGB550-E718 expression respect to the total cellular proteins expression. Specifically, we evaluated the interest protein expression using two widely used E. coli strains (BL21(DE3) and BL21-RP) to express heterologous proteins in the cytoplasm [21]. Once the E. coli strain was chosen, we designed a culture medium assay to improve the CIGB550-E718 expression. The expression levels of this recombinant protein in Bl21(DE3) E. coli, as well as its easy and efficient purification, make this fusion protein promising in terms of production and scale-up. Yields of the non-optimized process achieved by us, are comparable with those obtained by Granadillo and co-workers [20] based on the same technological platform. Besides, our results agree with those reported by other researchers, who have obtained the recombinant protein E7 for therapeutic use from both, HPV-18 and HPV-16, using the same E. coli Bl21(DE3) expression strain and obtaining similar levels of protein expression [22, 23]. It is known that the state of aggregation can be an important element for increasing the immunogenicity of recombinant proteins. Moreover, evidences show that particulate antigens play an important role in generating a CTL immune response [24]. In this sense, the immunoidentification revealed aggregates of the recombinant protein above 21 kDa that could enhance the cellular immune response against the E7 antigen. The aggregation state of this molecule can be potentiated by the presence of a zinc binding domain composed by two cysteine motifs, which could be involved in the CIGB550-E718 protein oligomerization [25]. The monomer of the CIGB550-E718 migrates as a protein of 20 kDa which is greater than its theoretical MW of 15.9 kDa. This anomalous electrophoretic behavior in electrophoresis gel is in agreement with the reported characteristics of E7 protein [26]. It is known that E7 protein migrates with the same aforementioned electrophoretic pattern due its high acidic aminoacid residues [27, 28].
Nowadays, the development of new products through cheaper methods that allow to obtain accessible products for the poorest population, which report the highest incidences of cervical cancer, represents a constant need. In order to this, the use of E. coli as expression system is a well-studied and cost-effective means for obtaining recombinant proteins in cell cytoplasm [29]. The novel candidate proposed in this study was obtained using E. coli BL21(DE3) strain as host, according to the described benefits of this bacteria for vaccine production. Taking advantage of this expression system that we have used before with HPV-16 E7 protein, we guarantee a low-cost process because the strain can grow vigorously in minimal media without any expensive supplement [20]. This fact could translate into inexpensive therapeutic options for women living in low-income countries.
Upcoming works will consist of testing the biological activity of the CIGB550-E718. For this purpose, the methodologies used for the previous candidate based on the E7 of HPV-16 will be performed.
4. Conclusions
This work reports on the non-optimized process of expression and purification of the novel CIGB550-E718, that could be used as therapeutic alternative for women presenting malignancies related to the HPV-18 infection. The whole process described here constitutes a favorable setting for the future development of this candidate with 90% purity through an inexpensive process. Making the vaccine production process inexpensive could make the vaccine more accessible to developing countries where the highest incidence rates are found. Indeed, the future combination of the CIGB550-E718 with our previous candidate (CIGB550-E7) against lesions associated to HPV-16, could be an ideal new vaccine for treating 70% of female population affected by both viruses.
5. Methods
5.1. Fusion protein expression vector (pABA plasmid)
The gene (E7-18m), that encodes a mutein of HPV-18 E7 protein, was assembled and amplified by two-step of PCR [30], using eight oligonucleotides with overlapping ends (Table 1). To obtain the mutant of the viral HPV-18 E7 antigen, we substituted two amino acids by glycine specifically the cysteine at position 27 and the glutamic acid at position 29 (C27 and E29 bases by G). In addition, codon optimization for protein expression in E. coli was done using the DNAWorks web site: http://helixweb.nih.gov/dnaworks. The plasmid pPEPE7M-7, described by Granadillo et al. [12], containing tryptophan promoter, ampicillin selection marker and His tag, was digested with HindIII and BamHI restriction enzymes and the DNA fragment encoding for the HPV-16 E7 protein was replaced by the E7-18m gene. Thus, the recombinant plasmid designated as pABA comprises the covalent fusion of the E7-18m gene to the CPP CIGB550, followed by six histidine (6xHis) residues. The pABA vector was transformed into the DH10B chemically competent E. coli cells. The purified DNA showing the expected restriction pattern using HindIII, BamHI, and PstI enzymes was sequenced.
|
Code |
Sequence 5’…3’ |
bp |
|
E7/HPV18-1 |
CCCAAGCTTCATGGTCCGAAAGCGACACTGCAGGATATTGTGTTGCA |
47 |
|
E7/HPV18-2 |
CCCAGCAGATCCACCGGAATCTCATTCTGCGGTTCCAGATGCAACACAATATCCTGCT |
58 |
|
E7/HPV18-3 |
CCGGTGGATCTGCTGGGCCACGGCCAGCTGAGCGATAGCGAAGAAGAAAATGATGAT |
57 |
|
E7/HPV18-4 |
CGACGCGCAGGCAGATGCTGATGATTCACGCCATCAATTTCATCATTTTCTTCTTCGCA |
59 |
|
E7/HPV18-5 |
TCTGCCTGCGCGTCGTGCGGAACCCCAACGTCATACCATGCTGTGCATGTGCTGCAAGTG |
60 |
|
E7/HPV18-6 |
TCATCCGCGCTGCTTTCCACCACCAGTTCAATACGCGCTTCGCACTTGCAGCACATGCAC |
60 |
|
E7/HPV18-7 |
GAAAGCAGCGCGGATGATCTGCGTGCGTTTCAGCAGCTGTTTCTGAATACCCTGAGCTTT |
60 |
|
E7/HPV18-8 |
GCGGATCCCTGCTGGCTCGCGCACCACGGGCACACAAAGCTCAGGGTATTCAGAAACA |
58 |
|
bp: bases pairs |
||
Table 1: Oligonucleotides used to obtain the HPV-18 E7 mutein.
5.2. Expression of the recombinant protein CIGB550-E718
The chemically competent E. coli BL21(DE3) and BL21-RP strains were transformed with the pABA plasmid, pM240 (vector without the gene of interest) and pPEPE7M-7 vector as positive control [12]. The protein expression was evaluated in 16 culture media of different composition containing either glucose or glycerol as carbon sources. E. coli cells transformed with the aforementioned vectors were inoculated in 50 mL of LB medium containing ampicillin (100 μg/mL) and L-trp repressor at the same concentration. After 6 h of incubation at 37°C and 300 rpm in a shaker, these cultures were used to inoculate 500 mL of LB medium containing ampicillin at the same concentration. After 4h, 3 β-indole-acrylic acid was added to a final concentration of 40 μg/mL and the culture was grown for another 16 h to express the recombinant protein. The strain in which was obtained the highest percentage of expression for the recombinant protein was selected to test 16 culture media with different composition at the scale of 50 mL (Table 2). As carbon source was used either 10 g/L glucose or glycerol. All of culture media contained M9 salt minimal medium (6.5 g/L Na2HPO4, 3 g/L KH2PO4, 5 g/L NaCl, 1 g/L NH4Cl) enriched with 0.1 M CaCl2, 1 M MgSO4 and 0.1 g/L ampicillin. Others components were added to supplement M9 salt medium, such as 5 g/L tryptone, 10 g/L casein hydrolysate, and 1 g/L yeast extract. Minimal media were evaluated alone and their combination with above-mentioned components as indicated in Table 2. The expression and purity of protein were evaluated by densitometry (TDI-1D manager 2.0 software, Spain) from 15% SDS-PAGE gels. Proteins were visualized by Coomasie blue staining.
|
Variants |
M9 salt minimal medium (M9) |
Casein hydrolysate (HC) |
Yeast extract (YE) |
Tryptone (T) |
CaCl2, MgSO4 and ampicillin |
|
A |
+ |
- |
- |
- |
+ |
|
B |
+ |
- |
+ |
- |
+ |
|
C |
+ |
- |
- |
+ |
+ |
|
D |
+ |
+ |
- |
- |
+ |
|
E |
+ |
- |
+ |
+ |
+ |
|
F |
+ |
+ |
+ |
- |
+ |
|
G |
+ |
+ |
- |
+ |
+ |
|
H |
+ |
+ |
+ |
+ |
+ |
|
The presence or absence of component in culture medium has been indicated by (+) or (–), respectively. Variants from A to H were used with glycerol or glucose as a carbon source. |
|||||
Table 2: Variants of culture media used for protein expression assay.
5.3. Purification of the recombinant protein CIGB550-E718
The purification of the CIGB550-E718 was carried out by a single immobilized metal affinity chromatography (IMAC) followed by a molecular exclusion step (G25) for buffer changing. This process was performed following Granadillo’s protocol [20] with some modifications. Specifically, we carried out cell disruption of 10 g biomass in French Press (Othake, Japan) at 1500 bar, with four passes at 4°C. The following steps were maintained. Samples in each step of the process were collected and later analyzed by SDS-PAGE and Western blot.
5.4. SDS-PAGE and Western Blot
SDS-PAGE (15%) was performed according to Laemmli [31] and the identity of the protein was confirmed by Western blot [32]. Briefly, 20 µL of each sample were applied in electrophoresis gel at 15%. After proteins separation, samples were transferred to Hybond C extra nitrocellulose (Amersham Biosciences) in a semidry transfer unit (Bio-Rad) for western blot analysis. The membrane was then blocked with a blocking solution consisting in 5% skimmed milk in PBS (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4 and 1.8 mM KH2PO4, pH 7.4)/0.1% Tween 20. After washing three times with 0.1% PBS-Tween 20, membrane was incubated with a goat anti-HPV-18 E7 polyclonal antibody (Abcam) diluted 1:50 for 2 h at 25°C. Subsequently to washed step, membrane was incubated with an anti-goat IgG antibody conjugated with HRP (Sigma) diluted 1:5000 for 1 h at the same temperature. The recognized bands were then visualized using a chemiluminescence kit (Amersham Pharmacia Biotech) after repetitive washes. All antibody dilutions were prepared with blocking solution.
6. Statistical analysis
GraphPad Prism software version 6 was used for the statistical analysis. In each case, normality was confirmed by Shapiro-Wilk test and the homogeneity of variance using Levene´s test. The evaluation of differences between culture media, were analyzed by the nonparametric Kruskal-Wallis test followed by a Dunn’s test. A value of alpha equal to 0.05 was prefixed as significance level.
Declarations
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing interests
None
Funding
BioCubaFarma/Center for Genetic Engineering and Biotechnology (CIGB)/IBM
Authors' contributions
IT, GM and RN conceive of the study, RN wrote and edit the manuscript while IT and GM reviewed it before submission. IT designs the sequence of the E7 mutein. BA, AA and RN carried out the fermentation process and purification of the fusion protein. Statistical analysis and figures were done by RN. All authors critically reviewed the manuscript and agreed to the published version of the manuscript.
Acknowledgements
We thank Estela D. Alvarez and Ariana Garcia Ojalvo for English revision. The research was supported by the Center for Genetic Engineering and Biotechnology.
Authors' information
Not applicable
References
- Arbyn M, Weiderpass E, Bruni L, et al. Estimates of incidence and mortality of cervical cancer in 2018: a worldwide analysis. The Lancet Global Health 8 (2020): e191-e203.
- Samimi G, Trabert B, Duggan MA, et al. Processing of fallopian tube, ovary, and endometrial surgical pathology specimens: a survey of US laboratory practices. Gynecologic oncology 148 (2018): 515-520.
- Demarco M, Hyun N, Carter-Pokras O, et al. A study of type-specific HPV natural history and implications for contemporary cervical cancer screening programs. EClinicalMedicine 22 (2020): 100293.
- Cheng L, Wang Y, Du J. Human papillomavirus vaccines: An updated review. Vaccines 8 (2020): 391.
- LaVigne AW, Triedman SA, Randall TC, et al. Cervical cancer in low and middle income countries: addressing barriers to radiotherapy delivery. Gynecologic oncology reports 22 (2017):16-20.
- Rybicki EP. Plant-based vaccines against viruses. Virology journal 11 (2014): 1-20.
- Li G, Sturgis EM. The role of human papillomavirus in squamous carcinoma of the head and neck. Current oncology reports 8 (2006): 130-139.
- Jabbar B, Rafique S, Salo-Ahen OM, et al. Antigenic peptide prediction from E6 and E7 oncoproteins of HPV types 16 and 18 for therapeutic vaccine design using immunoinformatics and MD simulation analysis. Frontiers in immunology 9 (2018): 3000.
- Ma B, Maraj B, Tran NP, et al. Emerging human papillomavirus vaccines. Expert opinion on emerging drugs 17(2012): 469-492.
- Yang A, Jeang J, Cheng K, et al. Current state in the development of candidate therapeutic HPV vaccines. Expert review of vaccines 15(2016): 989-1007.
- Lin K, Doolan K, Hung C-F, et al. Perspectives for preventive and therapeutic HPV vaccines. Journal of the Formosan Medical Association 109 (2010): 4-24.
- Granadillo M, Vallespi MG, Batte A, et al. A novel fusion protein-based vaccine comprising a cell penetrating and immunostimulatory peptide linked to human papillomavirus (HPV) type 16 E7 antigen generates potent immunologic and anti-tumor responses in mice. Vaccine 29 (2011): 920-930.
- Stirdivant SM, Ahern JD, Oliff A, et al. Retinoblastoma protein binding properties are dependent on 4 cysteine residues in the protein binding pocket. Journal of Biological Chemistry 267 (1992): 14846-14851.
- Lee J-O, Russo AA, Pavletich NP. Structure of the retinoblastoma tumour-suppressor pocket domain bound to a peptide from HPV E7. Nature 391 (1998): 859-865.
- Chan HM, Smith L, La Thangue NB. Role of LXCXE motif-dependent interactions in the activity of the retinoblastoma protein. Oncogene 20 (2001): 6152-6163.
- Helt A-M, Galloway DA. Mechanisms by which DNA tumor virus oncoproteins target the Rb family of pocket proteins. Carcinogenesis 24 (2003): 159-169.
- Granadillo M, Torrens I, Guerra M, et al. A novel strategy to improve antigen presentation for active immunotherapy in cancer. Fusion of the human papillomavirus type 16 E7 antigen to a cell penetrating peptide. Biotecnología Aplicada 29 (2012): 194-197.
- Yang J, Luo Y, Shibu MA, et al. Cell-penetrating peptides: Efficient vectors for vaccine delivery. Current drug delivery 16 (2019): 430-443.
- Vallespi MG, Glaria LA, Reyes O, et al. A Limulus antilipopolysaccharide factor-derived peptide exhibits a new immunological activity with potential applicability in infectious diseases. Clinical and diagnostic laboratory immunolog 7 (2000): 669-675.
- Granadillo M, Batte A, Lugo VM, et al. Expression, purification and characterization of a recombinant fusion protein based on the human papillomavirus-16 E7 antigen. Springerplus 2 (2013): 1-8.
- Sørensen HP, Mortensen KK. Advanced genetic strategies for recombinant protein expression in Escherichia coli. Journal of biotechnology 115 (2005): 113-128.
- Fiedler M, Campo-Fernández B, Laich A, et al. Purification and characterisation of the E7 oncoproteins of the high-risk human papillomavirus types 16 and 18. Journal of virological methods 134 (2006): 30-35.
- Liu B, Ye D, Song X, et al. A novel therapeutic fusion protein vaccine by two different families of heat shock proteins linked with HPV16 E7 generates potent antitumor immunity and antiangiogenesis. Vaccine 26 (2008): 1387-1396.
- Snapper CM. Distinct immunologic properties of soluble versus particulate antigens. Frontiers in immunology 9 (2018): 598.
- Barbosa MS, Lowy DR, Schiller JT. Papillomavirus polypeptides E6 and E7 are zinc-binding proteins. Journal of virology 63 (1989): 1404-1407.
- Sang B-C, Barbosa MS. Single amino acid substitutions in" low-risk" human papillomavirus (HPV) type 6 E7 protein enhance features characteristic of the" high-risk" HPV E7 oncoproteins. Proceedings of the National Academy of Sciences 89 (1992): 8063-8067.
- Armstrong DJ, Roman A. The anomalous electrophoretic behavior of the human papillomavirus type 16 E7 protein is due to the high content of acidic amino acid residues. Biochemical and biophysical research communications 192 (1993): 1380-1387.
- Bolhassani A, Zahedifard F, Taghikhani M, et al. Enhanced immunogenicity of HPV16E7 accompanied by Gp96 as an adjuvant in two vaccination strategies. Vaccine 26 (2008): 3362-3370.
- Rosano GL, Ceccarelli EA. Recombinant protein expression in Escherichia coli: advances and challenges. Frontiers in microbiology 5 (2014): 172.
- Xiong A-S, Yao Q-H, Peng R-H, et al. A simple, rapid, high-fidelity and cost-effective PCR-based two-step DNA synthesis method for long gene sequences. Nucleic acids research 32 (2004): e98-e99.
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 (1970): 680-685.
- Burnette WN. “Western blotting”: electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Analytical biochemistry 112 (1981): 195-203.


 Impact Factor: * 4.1
Impact Factor: * 4.1 Acceptance Rate: 74.74%
Acceptance Rate: 74.74%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 