Expression of BRCA1 and BRCA2 mRNA in Ovarian Cancer Patients Attending a Tertiary Level Cancer Hospital in Bangladesh
Latifa Nishat1, Sufi Hannan Zulfiqar Rahman2, Farida Arjuman3, Mahenaz Afroz4, Rahinur Ara Rimon5, Nargis Sultana5, Sadika Sharmin5, Shafayat Mohammad Imteaz6
1Department of Anatomy, Bangladesh Medical University, Dhaka, Bangladesh
2Department of Immunology and Molecular Biology, National Institute of Cancer Research and Hospital, Dhaka, Bangladesh
3Department of Histopathology, National Institute of Cancer Research and Hospital, Dhaka, Bangladesh
4Department of Gynecological Oncology, National Institute of Cancer Research and Hospital, Dhaka, Bangladesh
5Department of Anatomy, Bangladesh Medical University, Dhaka, Bangladesh
6Trust Grade Registrar- CT level, Cumberland Infirmary, Carlisle, Cumbria, UK
*Corresponding Author: Dr. Latifa Nishat, Associate Professor, Department of Anatomy, Bangladesh Medical University (BMU), Shahbag, Dhaka- 1000, Bangladesh
Received: 22 June 2025; Accepted: 03 July 2025; Published: 14 July 2025.
Article Information
Citation: Latifa Nishat, Sufi Hannan Zulfiqar Rahman, Farida Arjuman, Mahenaz Afroz, Rahinur Ara Rimon, Nargis Sultana, Sadika Sharmin, Shafayat Mohammad Imteaz. Expression of BRCA1 and BRCA2 mRNA in Ovarian Cancer Patients Attending a Tertiary Level Cancer Hospital in Bangladesh. Journal of Cancer Science and Clinical Therapeutics. 9 (2025): 102-111.
View / Download Pdf Share at FacebookAbstract
BRCA genes play a role in the pathogenesis and effectiveness of ovarian cancer treatments. Variations in the expression of these genes have been observed in ovarian cancer in different populations. The expression of BRCA genes in Bangladeshi patients with ovarian cancer remains unexplored. Data on BRCA1 and BRCA2 gene expression are important for advancing personalized medicine, improving the early detection of ovarian cancer in this population, and enhancing the global understanding of the molecular behavior of ovarian cancer. This study aimed to analyze BRCA1 and BRCA2 mRNA expression in cancerous and non-cancerous ovarian tissues from Bangladeshi women. Total RNA was extracted from formalin-fixed paraffin-embedded (FFPE) ovarian cancer tissue of 44 patients with ovarian cancer and non-cancerous tissue from 29 oophorectomized patients. Two-step real-time RT-PCR of mRNA was performed to amplify BRCA1 and BRCA2 genes and to calculate mRNA expression using the 2-ΔΔCt method. Ovarian cancer tissues displayed significantly higher expression of both BRCA1 and BRCA2 mRNA than in the non-cancerous ovarian tissues (median of BRCA1 3.24 versus 0.77, p < 0.001 and median of BRCA2 2.90 versus 0.39, p = 0.001). BRCA2 mRNA expression was significantly higher (p = 0.015) in stage I ovarian cancer. A positive correlation was observed between BRCA1 and BRCA2 expression (Spearman’s rho = 0.428, p = 0.004). The elevated and positively correlated BRCA1 and BRCA2 gene expression suggests a coordinated regulatory mechanism in the pathogenesis of ovarian cancer. Higher BRCA2 expression in early stage ovarian cancer highlights its potential as a biomarker for early detection.
Keywords
<p>BRCA1 mRNA expression; BRCA2 mRNA expression; Ovarian cancer; FFPE ovarian cancer tissue; Bangladeshi ovarian cancer patient.</p>
Article Details
Abbreviations: BRCA- Breast Cancer gene; BRCA1- Breast Cancer gene1; BRCA2- Breast Cancer gene2; GAPDH- Glyceraldehyde-3-Phosphate Dehydrogenase gene; FFPE- Formalin-Fixed Paraffin-Embedded; BMU- Bangladesh Medical University; BSMMU- Bangabandhu Sheikh Mujib Medical University; NICRH- National Institute of Cancer Research and Hospital; RNA- Ribonucleic acid; mRNA- messenger RNA; FIGO- International Federation of Gynecology and Obstetrics
1. Introduction
Ovarian cancer is the most lethal type of gynecological cancer. Globally, it is ranked as the seventh most common cancer in women, with approximately 384,000 new cases and 207,000 deaths by 2022 [1,2]. Late diagnosis, limited therapeutic options, and lack of effective biomarkers for monitoring treatment response contribute to the high fatality rate of ovarian cancer [3]. Cytoreductive surgery followed by platinum-based chemotherapy is performed in newly diagnosed patients [4]. Despite this treatment, approximately 70% of patients relapse within the subsequent three years [3]. Recurrent ovarian cancer is usually incurable despite multiple additional lines of treatment [5]. However, the exact cause of ovarian cancer remains unknown. Lifestyle factors such as obesity, smoking, and unhealthy diet increase this risk [6]. The risk of occurrence also increases with family history of ovarian or breast cancer [7]. Mutations in the BRCA (BRCA1 and BRCA2) genes contribute to the development of hereditary and sporadic ovarian cancers [8,9]. BRCA1 or BRCA2 mutation carriers develop hereditary ovarian cancers approximately 10 times more than the non-carriers [10]. These genes are involved in cellular growth inhibition, apoptosis, and DNA damage repair via homologous recombination [11]. BRCA1 is also responsible for non-homologous end-joining (NHEJ) repair [12]. Mutations in these genes influence the effectiveness of ovarian cancer treatments. Ovarian cancer patients with BRCA mutations have shown better survival with platinum-based chemotherapy and poly(ADP-ribose) polymerase inhibitor (PARPi) drugs [13,14]. Currently, PARPi drugs are approved for all platinum-sensitive ovarian cancer patients, regardless of BRCA mutation status [15]. However, the literature also provides contradictory results, such as worse survival in hereditary ovarian cancer or similar survival in BRCA mutation-associated sporadic epithelial ovarian cancer. These contradictory findings have inspired scientists to search for underlying factors beyond known mutations. The presence of rare DNA sequence variants of unknown significance (VUS), epigenetic modifications, and mutations deep within introns influences the molecular behavior and effectiveness of drugs used in ovarian cancer treatment [5]. Because all of these factors can affect transcription, the expression of BRCA1 and BRCA2 mRNA may explain the pathogenesis of ovarian cancer. Researchers suggest that the expression status of BRCA1 or BRCA2 mRNA in ovarian cancer patients could help select patients suitable for platinum-based chemotherapeutics and PARPi and could also predict prognosis [16]. Evidence of highly variable BRCA1 and BRCA2 expression has been found in ovarian cancer [5]. Some studies found it to be downregulated, whereas others found it to be upregulated. The expression status of BRCA genes also predicts the prognosis of ovarian cancer. Studies have found better overall survival with low BRCA1 expression and better progression-free survival with low BRCA2 expression [5,16].
Mutation in BRCA1 and BRCA2 are known to significantly increase the risk of ovarian cancer. Prevalence and mutation spectra vary across populations. Research tailored to the Bangladeshi population can reveal unique genetic profiles, enhancing the understanding of hereditary cancer risks specific to this demographic. There is a paucity of data on BRCA gene expression and mutations in patients with ovarian cancer from Bangladesh. Filling this gap can facilitate the development of region-specific genetic screening protocols and personalized treatment strategies. Understanding the mRNA expression levels of BRCA genes may serve as a potential biomarker for early diagnosis or risk assessment. Early detection can considerably improve the prognosis and survival rates of patients with ovarian cancer. Knowledge of BRCA expression can inform targeted therapies such as PARP inhibitors, which are more effective in patients with BRCA-related deficiencies. Tailored treatment approaches can improve patient outcomes and reduce the adverse effects. Ovarian cancer is a significant health burden in Bangladesh, as it is frequently reported in Bangladeshi women after breast and cervical cancers, with a high mortality rate [17]. Local research can inform public health policies, screening programs, and resource allocation, ultimately contributing to the better management and reduction of mortality. The data generated from Bangladesh can add to the global understanding of ovarian cancer genetics, emphasizing the importance of diverse genetic research to ensure equitable healthcare advances worldwide. Therefore, this study is crucial for advancing personalized medicine, improving early detection, and enhancing treatment strategies tailored to the Bangladeshi population, thereby contributing to better clinical outcomes and health policies. This study aimed to analyze the expression of BRCA1 and BRCA2 mRNA in formalin-fixed paraffin-embedded (FFPE) ovarian cancer tissue of Bangladeshi women with ovarian cancer in comparison to non-cancerous ovarian tissue that had undergone oophorectomy due to other gynecological pathologies.
2. Materials and methods
This cross-sectional study was conducted in the Department of Anatomy, Bangladesh Medical University (BMU) in collaboration with the Department of Histopathology and the Department of Immunology and Molecular Biology of the National Institute of Cancer Research and Hospital (NICRH), Dhaka. From the records of the Department of Histopathology of NICRH, voluntarily agreed 45 histologically diagnosed Bangladeshi ovarian cancer patients and 30 patients whose ovarian tissues were sent for histopathology due to gynecological diseases other than ovarian cancer and who did not have malignant cells in their ovaries between January 2023 and July 2024 were selected for this study. The patients were selected according to the availability of good quality FFPE tissue block with sufficient amount of ovarian cancer or non-cancerous tissue (assessed by a histopathologist). All of the above mentioned patients were invited and the voluntarily agreed patients were selected for the study. Their reproductive and cancer-related characteristics were recorded by interviewing the patients, searching hospital records, and histopathology reports using a structured data collection sheet. Exclusion criterion was presence or had a history of other cancer. Among the 45 ovarian cancer patients one patient was excluded because the quality and quantity of the extracted RNA was not satisfactory. The sociodemographic and reproductive characteristics of the participants are shown in Supplementary Table S1. FFPE ovarian cancer tissue blocks of patients with ovarian cancer and non-cancerous ovarian tissue blocks of patients without ovarian cancer were retrieved from the tissue archive. Four tissue sections, each 10 mm thick, were collected from each FFPE tissue block as cancerous and non-cancerous ovarian tissue samples. RNA was extracted from the tissue sample sections, reverse transcribed to cDNA, and amplified using real-time PCR to analyze the expression of BRCA1 and BRCA2 mRNA.
2.1 RNA extraction
RNA was extracted from FFPE tissue sections using the PureLink FFPE Total RNA Extraction Kit (Invitrogen, Thermo Fisher Scientific, USA) according to the manufacturer’s instructions. The quality and quantity of the RNA were checked photometrically using an Eppendorf Biophotometer D30 (Eppendorf AG, Germany). We identified RNA from 44 ovarian cancer and 29 non-cancerous ovarian tissue samples eligible for downstream applications. In these samples, the RNA concentration was ≥12 ng/mL and the 260/280 absorbance ratio was 1.82 to 2.1. Samples with high RNA concentrations were diluted to <50 ng/mL. The extracted RNA was stored at -80°C until further use.
2.2 Reverse transcription of mRNA
Reverse transcription of mRNA was performed, and cDNA was synthesized using the Verso cDNA synthesis kit (Applied BiosystemTM, Thermo Fisher Scientific, USA) according to the manufacturer’s protocol using the ProFlex PCR System thermal cycler (Thermo Fisher Scientific, USA). The PCR profile was set as follows: one cycle at 42°C for 60 min for reverse transcription, followed by one cycle at 95°C for 2 min for enzyme inactivation. A blend of RNA primers containing random hexamers and oligo (dT) primers at a 3:1 ratio was used. The RT enhancer in the kit was used to break down contaminant DNA and augment cDNA synthesis. The newly synthesized cDNA was stored at -80 °C until further use.
2.3 Amplification of cDNA by Real-Time PCR
PCR amplification for gene expression analysis was performed using commercially available ready-to-use TaqMan gene expression (GE) assay mix kits (Applied Biosystems, Thermo Fisher Scientific, USA) for BRCA1, BRCA2, and GAPDH. Each GE assay mix consisted of a pair of unlabeled PCR primers, a TaqMan probe with a dye-labeled 5´ end, and a minor groove binder (MGB) and non-fluorescent quencher (NFQ) on the 3´end. The specifications of the assay kits are as follows:
BRCA1 GE Assay Kit: Assay No. Hs01556193_m1, catalogue no. 4331182 (FAM-MGB)
BRCA2 GE Assay Kit: Assay No. Hs00609073_m1, catalogue no. 4331182(FAM-MGB)
GAPDH GE Assay Kit: Assay No. Hs02786624_g1, catalogue no. 4448489(VIC-MGB)
BRCA1-GAPDH and BRCA2-GAPDH multiplex relative quantification PCR was performed using a 7500 Fast Dx Real-Time PCR System (Applied Biosystems, Thermo Fisher Scientific, USA), where BRCA1 and BRCA2 were the target genes and GAPDH was the endogenous control gene. BRCA1-GAPDH and BRCA2-GAPDH multiplex PCR reaction mix was prepared using TaqMan Fast Advanced Master Mix (Applied BiosystemTM, Thermo Fisher Scientific, USA), BRCA1/BRCA2 GE assay mix, GAPDH GE assay mix, cDNA, and nuclease-free water. The proportions of the reaction mix components are listed in Supplementary Table S2.
The PCR reaction profile was set as one cycle at 95°C for 20 sec for polymerase activation, followed by 45 cycles of denaturation at 95°C for 3 sec, primer annealing, probe hybridization, and extension at 60°C for 30 sec. The cycle threshold (Ct) values of BRCA1(FAM) and GAPDH(VIC) for each sample were obtained from the BRCA1-GAPDH multiplex PCR amplification curve, and BRCA2(FAM) and GAPDH(VIC) for each sample were obtained from the BRCA2-GAPDH multiplex PCR amplification curves. BRCA1, BRCA2, and GAPDH mRNA were amplified in all 44 cancer and 29 non-cancerous tissue samples.
2.4 Calculation of BRCA1/BRCA2 mRNA expression
The 2˗ΔΔCt of BRCA1 and BRCA2 mRNA expression was calculated from the Ct values of the BRCA1, BRCA2, and GAPDH amplification curves of cancerous and non-cancerous ovarian tissues using Microsoft Office Excel 2007.
2.5 Ethical issues
This research was conducted after obtaining approval from the institutional review board of BMU and NICRH. This study was conducted in accordance with the Declaration of Helsinki. All the participants were treated equally with respect to each other. Written informed consent was obtained from each participant after explaining the aim and possible benefits of the study in cancer research. Each participant was assigned a code number for anonymity. Safeguards were followed to ensure the confidentiality and security of the information. FFPE ovarian cancer and non-cancerous ovarian tissues were collected from the tissue archives. Therefore, there were no physical, psychological, social, or legal risks during sample collection.
2.6 Data analysis
The collected data were checked and edited manually for technical discrepancies. Histological classification of the tumors was performed according to the WHO classification system [18].The histological grade was assigned according to Matsune et al. [19]. Staging of ovarian cancer was performed according to the International Federation of Gynecology and Obstetrics (FIGO) staging system [20].
Statistical analyses were performed using the IBM SPSS Statistics version 20. The mean ± standard deviation (SD) was used for normally distributed data, and the median, interquartile range (IQR) was used for skewed data. The Shapiro-Wilk test was performed to analyze the distribution of data. The expression of BRCA1 and BRCA2 mRNA in ovarian cancer tissues was compared with that in non-cancerous tissues using the Mann-Whitney U test. The correlation between the expression of BRCA1 and BRCA2 genes was analyzed by Spearman rank test, and regression analysis was performed. Data were analyzed to determine the association between BRCA1 and BRCA2 mRNA expression and cancer-related characteristics using the Mann-Whitney U test for two groups and Kruskal-Wallis test for more than two groups. A p-value < 0.05 was considered statistically significant.
3. Results
3.1 Characteristics of ovarian cancer patients
The study involved patients with ovarian cancer diagnosed at a mean age of 44.73 ± 12.56 years. Most cases (86%) were epithelial in origin, predominantly of the high-grade serous ovarian cancer (HGSOC) subtype (78.95%). More than half of the patients (52%) were diagnosed with FIGO stage I. Most cancers were sporadic (86.40%), with some patients reporting a family history of cancer, including ovarian (1 patient), breast (2 patients), stomach, liver, and prostate cancers (3 patients). The cancer-related characteristics of the patients with ovarian cancer are presented in Table 1.
Table 1: Association of expression of BRCA1 and BRCA2 mRNA with cancer related characteristics of the patients (n = 44)
|
Cancer related characteristics |
Number (percentage) |
BRCA1 expression |
P value |
BRCA2expression |
P value |
|
Median (IQR) |
Median (IQR) |
||||
|
Mean age at diagnosis ± SD (year) 44.73 ± 12.56 |
|||||
|
Hereditary of cancer |
|||||
|
Hereditary |
6 (13.60) |
1.96 (1.90) |
0.078a |
1.88 (4.53) |
0.259a |
|
Sporadic |
38 (86.40) |
3.42 (3.20) |
3.32 (5.01) |
||
|
Cancer type |
|||||
|
Epithelial |
38 (86.40) |
3.24 (3.17) |
0.907a |
2.81 (4.86) |
0.493a |
|
Germ cell tumor |
6 (13.60) |
3.05 (4.69) |
4.82 (4.10) |
||
|
FIGO stage |
|||||
|
Stage I |
23 (52.30) |
3.22 (4.04) |
0.823b |
5.98 (8.36) |
0.015b |
|
Stage III |
17 (38.60) |
3.34 (2.75) |
2.17 (2.12) |
||
|
Stage IV |
4 (9.10) |
3.13 (8.37) |
2.30 (5.08) |
||
|
Grade (n = 38) |
|||||
|
Low grade |
8 (21.05) |
2.49 (9.12) |
0.368a |
4.18 (8.04) |
0.739a |
|
High grade |
30 (78.95) |
3.30 (2.70) |
2.81 (4.58) |
||
|
Histological subtype (n = 38) |
|||||
|
HGSOC |
28 (73.68) |
3.13 (2.78) |
0.216b |
2.64 (2.98) |
0.472b |
|
LGSOC |
2 (5.26) |
1.91 |
3.59 |
||
|
HGMOC |
2 (5.26) |
8.92 |
20.51 |
||
|
LGMOC |
6 (15.80) |
2.63 (13.58) |
4.34 (15.53) |
||
P value < 0.05 was considered as significant.
a, Mann-Whitney U test; b, Kruskal-Wallis test.
HGSOC, high-grade serous ovarian cancer; LGSOC, low-grade serous ovarian cancer; HGMOC, high-grade mucinous ovarian cancer; LGMOC, low-grade mucinous ovarian cancer.
BRCA1 and BRCA2 mRNA expression in ovarian tissue
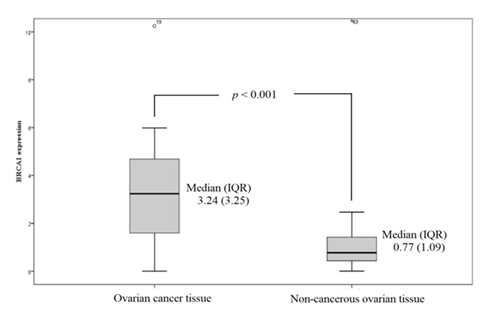
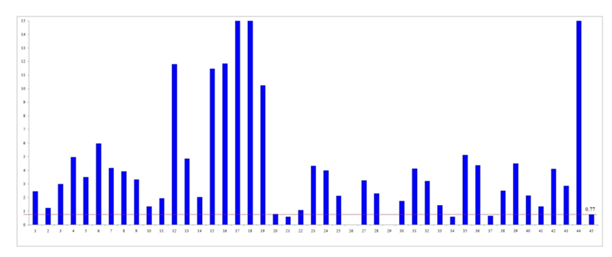
BRCA1: The median (IQR) of BRCA1 mRNA expression in cancerous and non-cancerous tissue was 3.24 (3.25) and 0.77 (1.09) respectively. The median BRCA1 mRNA expression level was significantly higher in cancerous tissues than in non-cancerous tissues (p < 0.001) (Figure 1). Most cancer tissues (except five samples) showed higher BRCA1 expression than the median of non-cancerous tissues, with six samples exhibiting more than 10-fold increased expression (Figure 2).
Figure 2: Expression level of BRCA1mRNA in individual ovarian cancer tissue in comparison to the median of the expression level of BRCA1 mRNA of non-cancerous ovarian tissues (bar 1 through bar 44 correspond to the sample number and bar 45 is the median of expression in non-cancerous ovarian tissue). BRCA1mRNA expression level of samples 17, 18 and 44 were 33.08, 30.88 and 29.71respectively which exceeded the limit of this graph.
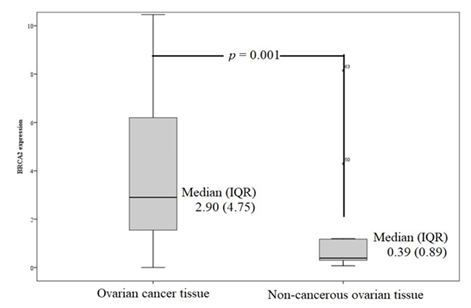
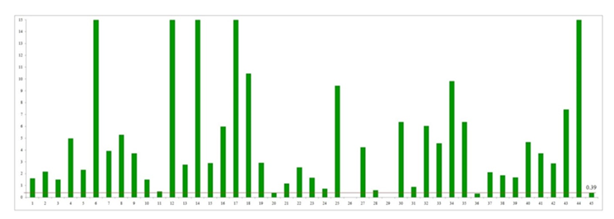
BRCA2: Similarly, BRCA2 mRNA expression was significantly higher in cancerous tissues than in non-cancerous ovarian tissues (p = 0.001). The median (IQR) of BRCA2 mRNA expression in cancerous and non-cancerous ovarian tissue was 2.90 (4.75) and 0.39 (0.89) respectively (Figure 3). All but two cancer samples had higher BRCA2 expression than the median of non-cancerous tissue, with 15 samples showing more than a 10-fold increase (Figure 4).
Figure 4: Expression level of BRCA2 mRNA in individual ovarian cancer tissue in comparison to the median of the expression level of BRCA2 mRNA of non-cancerous ovarian tissues (bar 1 through bar 44 correspond to the sample number, and bar 45 is the median of expression in non-cancerous ovarian tissue). BRCA2 mRNA expression levels of samples 6, 12, 14, 17, 18 and 44 were 35.04, 37.04, 109.40, 109.97 and 135.39 respectively which exceeded the limit of this graph.
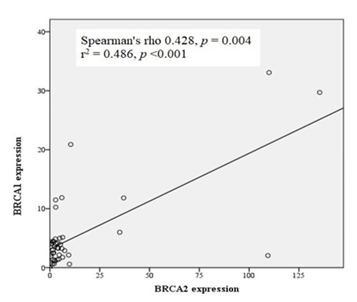
Correlation of expression: A fair but significant positive correlation was observed between BRCA1 and BRCA2 expression levels in cancer tissues (Spearman’s rho = 0.428, p = 0.004, r2 = 0.486, p < 0.001) (Figure 5).
Association with cancer-related characteristics: BRCA2 expression was significantly associated with FIGO stage; stage I cancers exhibited higher BRCA2 expression (median 5.98) than stages III and IV (median 2.17 and 2.30, respectively; p = 0.015). No significant associations were found between BRCA1 or BRCA2 expression levels and hereditary factors, histological type, grade, or subtype of ovarian cancer. The associations between BRCA1 and BRCA2 expression and cancer-related characteristics are presented in Table 1.
4. Discussion
The findings of this study revealed significant upregulation of both BRCA1 and BRCA2 mRNA expression in ovarian cancer tissues compared to non-cancerous ovarian tissues of Bangladeshi women. This suggested that these genes may contribute to ovarian tumorigenesis in this population. This result aligns with previous research suggesting that BRCA gene expression plays a critical role in ovarian tumor biology, although the direction of dysregulation (up- or down-regulation) has been inconsistent across studies. Custodio et al. found variable expression of BRCA1 and BRCA2 in 42 ovarian cancer FFPE tissue samples in Portugal; some tissues had comparable levels of expression, while some samples expressed 10 times higher levels than the normal fallopian tube FFPE tissue samples [5]. They found higher median expression of BRCA1 and BRCA2 mRNA in ovarian cancer tissue samples than in normal fallopian tissue. In another study in Austria, Tsibulak et al. found higher levels of BRCA1 and BRCA2 mRNA in 201 patients with ovarian cancer than in normal fallopian tissues [16]. Wang et al. analyzed The Cancer Genome Atlas (TCGA) datasets and found upregulation of both BRCA1 and BRCA2 in ovarian cancer. They also analyzed Oncomine data and found higher levels of BRCA1 and BRCA2 mRNA expression in ovarian cancer [21]. Tsibulak et al. suggested that the elevated transcriptional levels of BRCA1 and BRCA2 genes are the response of these genes to the high demand for homologous recombination repair in highly proliferating ovarian cancer cells [16]. BRCA1 and BRCA2 play a primary role in homologous recombination (HR) repair [11. The upregulation of BRCA gene expression depends on the cell cycle phase, and its peak level is obtained at the G1/S boundary [21]. Other factors associated with the upregulation of BRCA include somatic reversion mutations and copy number gain. Reversion mutations that restore wild-type amino acid sequences result in functional BRCA proteins [22,23]. Lheureux et al. found higher expression of BRCA genes in PARPi-resistant ovarian cancer due to the copy number gain of wild-type BRCA alleles [24]. BRCA1 and BRCA2 mRNA expression is altered by heterozygosity. Maxwell et al. identified differential inactivation of the normal allele of BRCA genes in ovarian cancer and breast cancer by loss of heterozygosity [25]. BRCA1 mRNA expression is inversely associated with BRCA1 DNA methylation status [16]. The upregulation observed in this study may reflect the unique genetic and molecular landscape of ovarian cancer in the Bangladeshi population, highlighting the importance of region-specific research.
In our study, BRCA1 and BRCA2 expression levels were lower in five and two samples, respectively, than the median expression in non-cancerous ovarian tissue, indicating the downregulation of this gene in a few cases. Studies have found lower expression of BRCA1 and BRCA2 due to epigenetic silencing by hypermethylation of CpG islands in the promoter region, mutations, and miRNA-mediated regulation [16,26]. The reduced expression of BRCA1 and BRCA2 mRNA observed in our study might be due to these factors. Further investigations are needed to elucidate the definitive etiology of the downregulation of these genes.
In this study, a positive correlation was observed between BRCA1 and BRCA2 expression. This finding aligns with the results of Egawa et al., who found a significant positive correlation between BRCA1 and BRCA2 mRNA expression in breast cancer in Japan [27]. Wang et al. and Jin et al. also observed overexpression of BRCA1 and BRCA2 in ovarian and breast cancer patients [21,28]. Custodio et al. also found coordinated expression of BRCA1, and BRCA2 genes with other 12 HR genes in Portugal [5]. This correlation indicates the coordinated regulation of these genes, which may synergistically influence DNA repair mechanisms or other pathways critical for tumor progression.
We found that BRCA2 expression was significantly higher in FIGO stage I cancers than in advanced-stage cancers. This suggests that BRCA2 overexpression may be an early event in tumorigenesis, offering a potential biomarker for early detection. Tsibulak et al. found a significant association between BRCA2 mRNA expression and tumor grade, and an association of both BRCA1 and BRCA2 expression with ovarian cancer types and histological subtypes, but not with FIGO stages [16]. We did not find any statistical association between the expression of BRCA1 or BRCA2 mRNA and ovarian cancer type, tumor grade, or histological subtype. In our study, most ovarian cancers were epithelial, compared to germ cell tumors (38 vs. 6). HGSOC was the predominant subtype (28 out of 38), and almost all were high-grade (30/38); a small number of other cancer types, histological subtypes, and low-grade cancers may be the reason for not getting any association, or it may be different in this population.
No significant differences in BRCA1 and BRCA2 expression were observed between hereditary and sporadic cancers. This implies that mRNA expression levels may not directly correlate with the germline mutation status. This underscores the complexity of BRCA gene regulation, which may involve epigenetic or posttranscriptional modifications.
The findings of this study may contribute to clinical decision-making. The upregulation of BRCA1/2 mRNA can influence the response to PARP inhibitors (PARPi) or platinum-based chemotherapy, as these therapies exploit DNA repair deficiencies. However, the paradoxical overexpression (rather than loss) observed in this study warrants further investigation into functional protein activity and potential resistance mechanisms. The association of high BRCA2 expression with early stage disease suggests its utility as a diagnostic or prognostic marker, particularly in resource-limited settings, such as Bangladesh, where advanced diagnostics are scarce.
Limitations
In this study, the predominance of high-grade serous ovarian cancer HGSOC (78.95%) limits the generalizability to other subtypes. Larger studies with more diverse histological types are needed. Another limitation was that the patients were not followed up. The absence of clinical outcome data precludes assessment of the prognostic value of BRCA1 and BRCA2 expression for survival or treatment response.
Conclusion
This study demonstrated that both BRCA1 and BRCA2 mRNA expression is significantly upregulated in ovarian cancer tissues compared to non-cancerous ovarian tissues, suggesting their potential role in ovarian tumor biology in the Bangladeshi population. The positive correlation between BRCA1 and BRCA2 expression indicates coordinated regulation, which may influence tumor development. Notably, higher BRCA2 expression in early stage (FIGO stage I) ovarian cancers suggests its potential utility as a biomarker for early detection. However, further research is needed to elucidate the functional implications of these expression patterns and their association with clinical outcomes, which could inform targeted therapeutic strategies and risk assessments in this population.
Acknowledgment
We are grateful to the patients who participated in this study. We thank the authority of Bangladesh Medical University for the funding support.
Author contributions
Conception or design of the work; or the acquisition, analysis, or interpretation of data for the work: LN, SHZR, FA, MA, RAR, NS, SS, SMI. Drafting the work or critically reviewing it for important intellectual content: LN, SHZR, FA, MA, RAR, NS, SS, and SMI. Final approval of the version to be published: LN, SHZR, FA, MA, RAR, NS, SS, and SMI. Accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved: LN, SHZR, FA, and MA.
Funding
This study was funded by the Research Grant for Teachers, Bangladesh Medical University (BMU) [former Bangabandhu Sheikh Mujib Medical University (BSMMU)], Dhaka, Bangladesh, Ref no. BSMMU/2023/13314(104).
Competing interests
The authors declare no conflict of interest.
Ethical approval
The study was approved by the Institutional Review Board of BMU (former BSMMU) [Ref no. BSMMU/2023/2246], and NICRH [Memo No. NICRH/IRB/2024/161].
Data availability statement
We confirm that the data supporting the findings of this study will be shared upon reasonable request.
Supplementary file
The sociodemographic and reproductive characteristics of patients with ovarian cancer and non-cancerous females are presented in Supplementary TableS1. The components and proportions of the PCR mix are listed in Supplementary Table S2. The calculation of 2˗ΔΔCt of BRCA1 and BRCA2 mRNA expression is included in the supplementary file.
References
- Bray F, Laversanne M, Sung H, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 74 (2024): 229-263.
- Webb PM, Jordan SJ. Global epidemiology of epithelial ovarian cancer. Nat Rev Clin Oncol 21(2024): 389-400.
- Lheureux S, Braunstein M, Oza AM. Epithelial ovarian cancer: Evolution of management in the era of precision medicine. CA Cancer J Clin 69 (2019): 280-304.
- Cortez AJ, Tudrej P, Kujawa KA, et al. Advances in ovarian cancer therapy. Cancer Chemother Pharmacol [Internet] 81 (2018): 17-38.
- Custódio N, Savisaar R, Carvalho C, et al. Expression Profiling in Ovarian Cancer Reveals Coordinated Regulation of BRCA1/2 and Homologous Recombination Genes. Biomedicines 10 (2022): 1-17.
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 70 (2020): 7-30.
- Kurman RJ, Shih IM. The dualistic model of ovarian carcinogenesis revisited, revised, and expanded. Am J Pathol [Internet] 186 (2016): 733-747.
- Zamwar UM, Anjankar AP. Aetiology, Epidemiology, Histopathology, Classification, Detailed Evaluation, and Treatment of Ovarian Cancer. Cureus 14 (2022).
- Kast K, Rhiem K, Wappenschmidt B, et al. German Consortium for Hereditary Breast and Ovarian Cancer (GC-HBOC). Prevalence of BRCA1/2 germline mutations in 21 401 families with breast and ovarian cancer. J Med Genet 53 (2016): 465-471.
- Grzymski JJ, Elhanan G, Morales Rosado JA, et al. Population genetic screening efficiently identifies carriers of autosomal dominant diseases. Nat Med 26 (2020): 1235-1239.
- Roy R, Chun J PS. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer 23 (2011): 68-78.
- Gorodetska I, Kozeretska I, Dubrovska A. BRCA genes: The role in genome stability, cancer stemness and therapy resistance. J Cancer 10 (2019): 2109-2127.
- Harter P, Johnson T, Berton-Rigaud D, et al. BRCA1/2 mutations associated with progression-free survival in ovarian cancer patients in the AGO-OVAR 16 study. Gynecol Oncol [Internet] 140 (2016): 443-449.
- Ledermann JA. PARP inhibitors in ovarian cancer. Ann Oncol [Internet] 27 (2016): 40-44.
- Mateo J, Lord CJ, Serra V, et al. A decade of clinical development of PARP inhibitors in perspective. Ann Oncol 30 (2019): 1437-1447.
- Tsibulak I, Wieser V, Degasper C, et al. BRCA1 and BRCA2 mRNA-expression prove to be of clinical impact in ovarian cancer. Br J Cancer [Internet] 119 (2018): 683-692.
- Hospital Cancer Registry Report (2018-2020). National Institute of Cancer Research and Hospital, Dhaka (2022).
- Christina IM, Philipp F, Hauptmann S, et al. The new WHO classification of ovarian , fallopian tube , and primary peritoneal cancer and its clinical implications. Arch Gynecol Obstet 293 (2016): 695-700.
- Matsuno RK, Sherman ME, Visvanathan K, et al. Agreement for tumor grade of ovarian carcinoma: analysis of archival tissues from the surveillance, epidemiology, and end results residual tissue repository. Cancer Causes Control 24 (2013): 749-757.
- Prat J, Belhadj H, Berek J, et al. Figo’s staging classification for cancer of the ovary, fallopian tube, and peritoneum: Abridged republication. J Gynecol Oncol 26 (2015): 87-89.
- Wang Z, Zhang J, Zhang Y, et al. Expression and mutations of BRCA in breast cancer and ovarian cancer: Evidence from bioinformatics analyses. Int J Mol Med 42 (2018): 3542-3550.
- Sorrells S, McKinnon KE, McBratney A, et al. Longitudinal and multi-tissue molecular diagnostics track somatic BRCA2 reversion mutations that correct the open reading frame of germline alteration upon clinical relapse. npj Genomic Med [Internet] 6 (2021).
- Murciano-Goroff YR, Schram AM, Rosen EY, et al. Reversion mutations in germline BRCA1/2-mutant tumors reveal a BRCA-mediated phenotype in non-canonical histologies. Nat Commun 13 (2022): 1-10.
- Lheureux S, Bruce JP, Burnier J V, et al. Somatic BRCA1/2 recovery as a resistance mechanism after exceptional response to poly (ADP-ribose) polymerase inhibition. J Clin Oncol 35 (2017): 1240-1249.
- Maxwell KN, Wubbenhorst B, Wenz BM, et al. BRCA locus-specific loss of heterozygosity in germline BRCA1 and BRCA2 Nat Commun [Internet] 8 (2017): 1-11.
- He W, Zhu H, Zhang S, et al. Promoter Methylation Changes in DNA Damage-Response Genes in Ovarian Cancer and Their Correlation with Prognosis. Clin Exp Obstet Gynecol 51 (2024).
- Egawa C, Miyoshi Y, Taguchi T, et al. Quantitative analysis of BRCA1 and BRCA2 mRNA expression in sporadic breast carcinomas and its relationship with clinicopathological characteristics. Japanese J Cancer Res 92 (2001): 624-630.
- Jin TY, Park KS, Nam SE, et al. BRCA1/2 Serves as a Biomarker for Poor Prognosis in Breast Carcinoma. Int J Mol Sci 23 (2022): 1-14.
Supplementary text
Calculation of BRCA1/BRCA2 mRNA expression
The 2˗ΔΔCt of BRCA1 and BRCA2 mRNA expression was assessed from the Ct value of the BRCA1, BRCA2, and GAPDH amplification curves using Microsoft Office Excel 2007. First, the ΔCt of BRCA1/BRCA2 of for each sample was calculated using the formula: ΔCt = Ct of BRCA1/BRCA2 - Ct of GAPDH. Then, the ΔΔCt of BRCA1/BRCA2 of each sample was calculated using the formula: ΔΔCt of BRCA1/BRCA2 = ΔCt of BRCA1/BRCA2 of the selected sample – average ΔCt of BRCA1/BRCA2 of the non-cancerous (control) tissue. The BRCA1/BRCA2 gene expression in a given sample was represented as 2˗ΔΔCt of BRCA1/BRCA2.
Supplementary Table S1: Socio-demographic and reproductive characteristics of the participants
|
Reproductive characteristic |
Ovarian cancer patient |
Non-cancerous female |
p-value |
|
(n = 44) |
(n = 30) |
||
|
Mean age (year) ± SD |
45.16 ± 12.37 |
48.43 ± 12.13 |
0.35 |
|
Median BMI (IQR) (kg/m2) |
21.67 (3.35) |
23.39 (3.55) |
0.004 |
|
Education, n(%)a |
|||
|
Primary |
9 (20.47) |
11 (36.67) |
|
|
Secondary and above |
16 (36.36) |
8 (26.66) |
0.295 |
|
No formal education |
19 (43.18) |
11 (36.67) |
|
|
Median (IQR) age at menarche (year) |
12 (3) |
12 (3) |
0.356 |
|
Menstrual cycle, n(%)b |
|||
|
Regular |
43 (97.73) |
23 (76.67) |
0.006 |
|
Irregular |
1 (2.27) |
7 (23.33) |
|
|
Menstrual status, n (%)a |
|||
|
Menstruating |
23 (52.27) |
14 (46.67) |
0.636 |
|
Postmenopausal |
21 (47.73) |
16 (53.33) |
|
|
Median (IQR) menopausal age (years) |
48 (5) |
48(4) |
0.718 |
|
Marital status, n (%)a |
|||
|
Married |
42 (95.45) |
28 (93.33) |
0.23 |
|
Unmarried and others |
2 (4.55)) |
2 (6.67) |
|
|
Median (IQR) number of children |
2 (2) |
2 (1) |
0.95 |
|
Contraceptive use, n (%)a |
|||
|
Never used |
28 (63.64) |
16 (53.33) |
|
|
Used |
16 (36.36) |
14 (46.67) |
0.657 |
P value < 0.05 was considered as significant
a, Chi-Square (χ2) test was performed; b, Fisher’s exact test was performed.
Unpaired t-tests and Mann-Whitney U tests were performed for numerical variables to determine the differences in means and medians, respectively.
Supplementary Table S2: Component and proportion of PCR reaction mix
|
Reaction component |
Volume (mL) |
|
|
BRCA1-GAPDH PCR |
BRCA2-GAPDH PCR |
|
|
TaqMan Fast Advanced Master mix |
10 |
10 |
|
BRCA1 GE assay mix |
1 |
0 |
|
BRCA2 GE assay mix |
0 |
1 |
|
GAPDH GE assay mix |
1 |
1 |
|
Nuclease free water |
4 |
4 |
|
cDNA template |
4 |
4 |
|
Reaction volume |
20 |
20 |


 Impact Factor: * 4.1
Impact Factor: * 4.1 Acceptance Rate: 74.74%
Acceptance Rate: 74.74%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 