MET-FISH Evaluation Algorithm: Proposal of a Simplified Method
Roberta Castiglione1,2, Christina Alidousty2, Barbara Holz2, Nicolai Duerbaum2, Maike Wittersheim3, Elke Binot2, Sabine Merkelbach-Bruse2, Nicolaus Friedrichs2, Matthias S Dettmer1, Alexander Bosse1, Reinhard Buettner2 and Anne Maria Schultheis2*
1Institute of Pathology, Klinikum Stuttgart, Stuttgart, Germany
2Institute of Pathology, University Hospital Cologne, Cologne, Germany
3Institute of Pathology, Medizin Campus Bodensee, Friedrichshafen, Germany
*Corresponding Author: Anne Maria Schultheis, Institute of Pathology, University Hospital Cologne, Cologne, Germany.
Received: 16 June 2022; Accepted: 05 September 2022; Published: 12 December 2022
Article Information
Citation: Roberta Castiglione, Christina Alidousty, Barbara Holz, Nicolai Duerbaum, Maike Wittersheim, Elke Binot, Sabine Merkelbach-Bruse, Nicolaus Friedrichs, Matthias S Dettmer, Alexander Bosse, Reinhard Buettner and Anne Maria Schultheis. MET-FISH Evaluation Algorithm: Proposal of a Simplified Method. Journal of Cancer Science and Clinical Therapeutics 6 (2022): 411-427.
View / Download Pdf Share at FacebookAbstract
MET amplifications (METamp) occur in 5% of NSCLC and represent in most case mechanisms of resistance to ALK and/or EGFRtargeted therapies. METamp detection can be performed using different techniques, although Fluorescence In-Situ Hybridization (FISH) remains the gold-standard, especially in the context of subclonality. To date current evaluation algorithms of MET amplifications are time consuming. Aim of the study was to identify a faster, equally reliable diagnostic algorithm for the detection of METamp, which is currently classified in negativity and low/intermediate/high-level amplification. N=497 NSCLC cases with available MET-FISH data had been selected. The results based on the first evaluated 20 cells had been re-calculated and compared with the definitive results based on 60 cells. For n=464 (93.4%) identical results had been obtained when counting 20 cells instead of 60 cells. Thirty-three cases (5.6%) showed a discrepancy, leading to an incorrect upgrade to a higher diagnostic category (n=25) and to an incorrect downgrade (n=8). We propose a simplified, yet equally reliable MET FISH-algorithm: after accurate screening of the whole tumor slide, twenty tumor cells have to be evaluated and results calculated: If the result is negative, or if all criteria of high-level METamp are fulfilled, the case can be signed out as such. All other cases should be considered as equivocal and additional 40 cells have to be counted. Given that, reliable results can be obtained by counting 20 cells only and an “equivocal” category for cases that need further investigation have been clearly defined.
Supplementary File
Keywords
<p>Evaluation Algorithm; Fish; MET-Amplification; Non-Small- Cell-Lung Cancer</p>
Article Details
1. Introduction
Lung cancer remains the main cause of cancer related deaths worldwide [1]. New therapies targeting specific genetic alterations fundamentally changed the treatment of patients with advanced stage disease [2]. As consequence, the identification of actionable genetic alterations is part of the routine diagnostic setting [3]. One of the most recent targets are aberrations of the Mesenchymal epithelial transition factor (MET) [4, 5]. According to current data [4], MET is altered in approximately 5% of non-small cell lung cancers (NSCLC) and is considered as adverse prognostic factor [6-11] and a potential predictive marker for response to anti MET therapeutics [12]. MET pathway activation occurs by several mechanisms that affect cancer cell survival, growth, and invasiveness [4, 12]; among those, MET amplifications. Preclinical and clinical evidence suggests that MET amplifications may occur as primary oncogenic driver in subsets of treatment-naive lung cancers as well as a secondary driver of acquired resistance after treatment with targeted therapies [12]. MET copy number gains arise from two distinct processes: polysomy and amplification [13]. While polysomy occurs in cases of chromosomal duplication, true amplification occurs in the setting of focal gene duplication and represents a real oncogenic driver [14, 15]. MET inhibition is now confirmed to lead to clinically meaningful antitumor activity with rapid and durable response and low side effects, especially in the context of MET exon 14 skipping mutations or higher level of amplifications [10, 16, 17].
- MET Amplification as Resistance Mechanism in EGFR- or ALK-altered NSCLC
MET activation has been implicated as an oncogenic driver in epidermal growth factor receptor (EGFR)- or anaplastic lymphoma kinase (ALK)-positive NSCLC and can mediate primary and secondary resistance to ALK and EGFR tyrosine kinase inhibitors (TKI) [18, 19]. MET inhibition may overcome ALK resistance combining MET and ALK inhibition [19, 20] or analogously, combining MET and anti-EGFR inhibition (erlotinib) [21] to overcome MET amplification mediated resistance to EGFR inhibition [22].
1.2 Identification of MET Amplification
The definition of clinically relevant MET amplification however has been a long-debated topic and several criteria have been proposed. MET amplification can be identified through different molecular techniques, however, the most widely used method in the clinical setting remains Fluorescence In-Situ Hybridization (FISH), also allowing the detection of the exact number of gene copies per cell. Numerous ongoing trials enrolling MET amplified NSCLC, show that cut-offs for the definition of MET amplification vary [22]. Recently, early data from clinical studies point out [12, 16] that a high-level amplification is necessary to obtain a significant clinical response [16]. As the level of amplification seems to be of prognostic [4, 23] and predictive value [5], FISH remains the most reliable, fast and cost-effective method for the identification of MET amplification in the clinical setting [4]. However a unique definition of high-level amplification is still debated. Some authors propose to use the MET/centromere 7 (CEP7) copy number ratio [16, 24, 25]. As described by Schildhaus et al [23], MET amplification can also co-occur in the context of amplification of the centromere, leading to a ratio < 2, so that the author has proposed a more complex approach including both MET gene copy number amplification and MET/centromere 7 copy number ratio. This approach has also been preferred in numerous large clinical trials (such as NCT02414139 NCT01610336, NCT01982955 (INSIGH study), and NCT02143466 (TATONN study)) [22]. In addition, in a very recent study [4] Overbeck et al propose a new category of MET high-level amplification identifying the higher unequivocal MET-amplification level, defined as average MET gene copy number of ≥ 10 independently from the MET/centromere 7 copy number ratio. With a steadily increasing number of cases, currently applied FISH-criteria can be time-consuming in the daily clinical setting and sometimes may not even be possible in the case of very small biopsies, not reaching the required number of tumor cells. Therefore, the aim of the current work was to develop a faster and equally reliable MET-FISH evaluation algorithm that allows the identification of all potentially relevant MET-amplification levels in basically all patient samples.
2. Materials and Methods
2.1 Case Collection
The archives of the Institute of Pathology at the University Hospital of Cologne, Cologne, Germany and of the Institute of Pathology at Klinikum Stuttgart, Stuttgart, Germany have been searched retrospectively for NSCLC cases at every stages, for which MET-FISH analysis has been performed as part of the routine diagnostic work-up according to the internal protocol of the Network Genomic Medicine established in Cologne. For the purpose of this study, only biopsy or resection specimens have been included in the study resulting in n=400 consecutive cases derived from the Institute of Pathology of the University Hospital of Cologne and n=97 consecutive cases from the Institute of Pathology at Klinikum Stuttgart. Cytology samples were excluded as specific cut-off criteria were established on formalin-fixed paraffin-embedded (FFPE) tissues samples. Of n=497 cases, n=400 (80.5%) were classified as negative, n=60 (12.1%) as low-level amplification, n=11 (2.2%) as intermediate-level amplification, n=26 (5.2%) as high-level amplification.
2.2 Histomorphological Analysis
Histology was reviewed by 3 experienced pathologists (AMS, RC and RB). The diagnosis of NSCLC was confirmed in all cases. Prior to the study, approval by the local ethics committee had been granted and all patients had signed written informed consent.
2.3 Fluorescence In-Situ Hybridization (FISH)
MET-FISH was performed as part of the routine diagnostic setting using a commercially available FISH probe Zytolight® SPEC MET/CN7 Dual color Probe (product nr. Z-2087-200) and its implementation kit (product nr. Z-2028-5/-20) provided by ZytoVision (ZytoVision GmbH, Bremerhaven, Germany) according to manufacturer’s instructions. The probe is composed by ZyGreen-marked (excitation 503 nm/ emission 528 nm) gene-sequence including the MET-region and by ZyOrange-marked (excitation 547 nm, emission 572 nm) gene-sequence including the centromeric alphasatellite-region D7Z1 of chromosome 7. Slides were reviewed at high magnification power (×63) and scored according to previous published guidelines [23]: the entire tumor area was initially screened and the spots with most signals were selected for further analysis. A total number of n=60 cells derived from 3 areas of 20 continuous cells each were counted. As previously described by our group, green MET signals and orange CEP7 were counted during routine diagnostic defining 4 groups of amplification status [23]:
- High-level amplification defined as tumors with MET/CEP7 ratio ≥ 2.0 or an average MET gene copy number per cell of ≥ 6.0 or ≥ 10% of tumor cells containing ≥ 15 MET signals.
- Intermediate-level of gene copy number (GCN) gain defined as ≥ 50% of cells containing ≥ 5 MET signals and criteria for high-level amplification not being fulfilled.
- Low-level of gene copy number gain defined as ≥ 40% of tumor cells showing ≥ 4 MET signals and criteria for high-level amplification or intermediate-level of gene copy number gain not being fulfilled.
- All other tumors were classified as negative.
An example of each category is depicted in Figure 1. In order to verify, whether a more effective evaluation algorithm was possible, the results of the original reports were pulled and re-calculated first using only the first counted 20 cells and then compared to the final result based on 60 counted cells. In both cases the same endpoints (MET mean GCN, CEP7 mean GCN, MET/CEP7 GCN-ratio and number of cells with ≥ 4/ ≥ 5/ ≥ 15 MET signals) were used and the cases were classified according to the following categories:
- Identical results when counting 20 and when counting 60 cells.
- Discrepant results leading to a potential upgrade to a higher category when counting only 20 cells.
- Discrepant results leading to a potential downgrade to a lower category when counting only 20 cells.
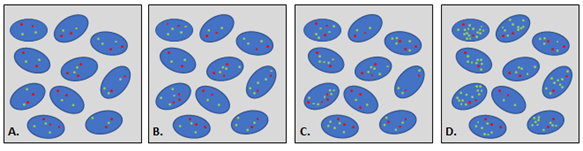
Figure 1: Different level of amplification. Red dot: copy of centromere 7, green dot: copy of MET gene. A. Negativity; B. Low-level amplification; C. intermediate-level amplification; D. high-level amplification.
2.4 Data Analysis
Automated evaluation of the number of signals of MET, CEP7 as well as calculation of MET/CEP7 ratio and identification of low- /intermediate- / high-level amplification were performed using the FileMakerÒ software and Microsoft Office Excel.
3. Results
The reported frequency of MET positivity was similar to the main literature data [4, 23]. The dataset of n=497 cases was distributed as follow: (i.) n=26 cases showed high-level amplification. Five of them would have been now classified as Top-level according to the new classification proposed by Overbeck and colleagues; (ii.) n=11 intermediate-level amplification. (iii.) n=60 low-level amplification; (iv.) n=400 negative result. Comparing the results after counting 20 or 60 cells, high concordance was observed. In details, for n=464 (93.4%) cases identical results were obtained when counting only 20 cells instead of 60 cells were obtained. N=25 cases (5.0%) cases would be upgraded to higher category when counting only 20 cells. N=8 cases (1.6%) would be downgraded counting only 20 cells (Figure 2 and Supplementary Table 1). The results obtained from our re-analysis lead to the proposal of the following diagnostic algorithm, which allows a more rapid, but equally reliable and precise evaluation of MET gene copy numbers (Figure 3). For all cases classified as negative, the analysis is completed after counting 20 representative cells and the case can be signed out as such. Cases classified as high-level amplification by fulfilling all three criteria (MET-GCN ≥ 6, at least 10% of tumor cells containing ≥ 15 MET signals and MET/CEP7 ratio ≥ 2) can be signed out as high-level amplification, otherwise (i.e. if only one or two of the criteria for high-level amplification are fulfilled) additional 40 cells have to be evaluated. For all cases that reach the criteria for low-level or intermediate-level amplification, the evaluation of additional 40 cells is mandatory. Given the current evidence of a significant prognostic value of top-level amplifications, defined by gene copy number higher then 10, we suggest including this new category as proposed by Schildhaus and colleagues [5].
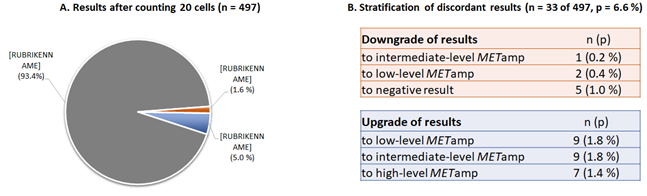
Figure 2: Comparison of results after counting 20 or 60 cells. A. Concordant results have obtained in n=464 (93.4%) of cases, discordant results in n=33 (6.6%) cases. B. Analysis of the n=33 discrepant cases after counting 20 or 60 cells. 8 cases (1,6%) would be downgraded in a lower diagnostic category after counting only 20 cells as following: n=5 (1.0%) downgraded to negative result instead of low-level amplification; n=2 (0.4%) downgraded to low-level instead of intermediate-level amplification; n=1 (0.2%) downgraded to intermediate-level instead of high-level amplification. 25 cases (5.0%) would be upgraded to higher category when counting only 20 cells: n=9 (1.8%) upgraded to low-level result instead on negative; n=9 (1.8%) upgraded to intermediate-level instead of low-level amplification; n=5 (0.4%) upgraded to high-level instead of low- or intermediate-level amplification.
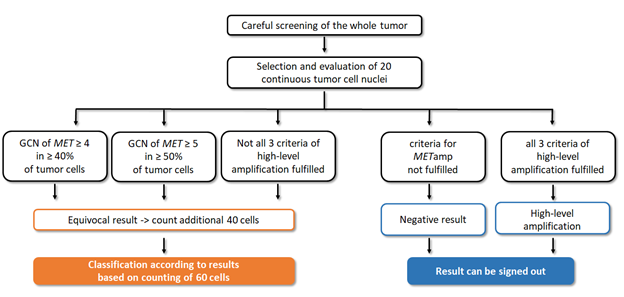
Figure 3: Diagnostic algorithm of MET amplification. After screening the whole tumor slide, the area with greater number of signals per cell is evaluated counting 20 cells. In case of preliminary low-level or intermediate-level amplification, or just one/two criteria for high-level amplification are fulfilled, 40 more cells need to be counted. The final result is based on the evaluation of 60 cells according to previous described criteria. In case of negative result as well as if all three criteria for high-level amplification are fulfilled, the analysis can be signed down.
4. Discussion and Conclusions
The identification of MET genetic alterations represents a crucial point in the molecular assessment of non-small cell lung cancer in advance stage of disease since MET may act both as primary driver and as mechanism of resistance to ALK-/EGFR- delivering therapies [12, 18, 19]. Over the last two years, the importance of MET alterations has further grown since two drugs targeting MET, capmatinib [5] and tepotinib [26], obtained FDA approval. With a steadily increasing number of cases of lung cancer needing a molecular characterization and the fact that FISH currently represents the gold-standard for detection of MET amplifications, MET FISH analyses can be time-consuming when using currently applied evaluation criteria. Clear cut-off criteria for MET amplification remain debated, but the most recent studies on MET amplifications agree that higher levels of amplification better respond to MET targeted therapies in clinical studies [5, 26] and are associated with poorer prognosis [4]. The use of next generation sequencing in the assessment of MET amplification, although time-sparing is not suggested since the sensibility in not superior and it provides no description concerning any subclonal variation [6]. The use of immunohistochemistry is as well not recommended for the identification of MET amplification, since the specificity is too low, as already published by our group [27]. In this work an updated, shorter, yet equally reliable algorithm for the evaluation of MET amplification has been presented, improving the current diagnostic algorithm [23]. According to the results based on n=497 unselected, consecutive MET FISH analyses of NSCLC, the same results are obtained in 93.4% (n=464) of cases when counting 20 cells instead of 60 cells. Of thirty-three (6.6%) discordant cases, n=25 (5.0%) would have been upgraded and n=8 (1.6%) would have been downgraded when counting only 20 cells, as listed in the results section and in Figure 2. An upgrade from negative result to low-level amplification (n=9, 1.8%) as well as an upgrade from low-level to intermediate-level amplification (n=9, 1.8%) can be avoided as cases are added to the “equivocal” category and need counting of additional 40 cells as described by the algorithm. Seven (n=7, 1.4%) cases would have been falsely classified as high-level when only counting 20 cells. However, this relevant misclassification can be avoided, if only cases that fulfil all three possible criteria for high-level amplification (MET/CEP7 ratio ≥ 2.0 and average MET gene copy number per cell of ≥ 6.0 and ≥ 10% of tumor cells containing ≥ 15 MET signals) are directly classified as high-level after counting 20 cells. All other cases are classified as “equivocal” and need the counting of additional 40 cells. Of note, this may lead to missing cases with very focal high-level amplification when thorough screening of a case is not performed. If a very focal high-level amplification in therapy-naïve patients however is of clinical relevance needs to be evaluated in further studies. In addition, we believe that the downgrade to a negative result instead of the identification of a clinically not relevant low-level amplification (n=5, 1.0%) can be prevented by thorough slide screening but is rather of no clinical relevance. All cases classifying as low-level (n=2, 0.4%) or intermediate-level (n=1, 0.2%) when using 20 cells will automatically be considered “equivocal” and will be reanalyzed, resulting in counting a total of 60 cells. In contrast to next-generation sequencing based methods, even if in our algorithm just 20 cells are counted, the clonal variations of amplifications are still not missed out, since before starting the count the whole tumor surface has to be screened looking for clonal events. In conclusion, here we propose an updated MET-FISH algorithm that provides a much more efficient yet equally reliable way to identify clinically relevant MET amplifications in NSCLC in the routine clinical setting, even in the case of very small biopsies.
Conflict of Interest
RB provided lectures and was part of Advisory Boards for AbbVie, Amgen, AstraZeneca, Bayer, BMS, Boehringer-Ingelheim, Illumina, Lilly, Merck-Serono, MSD, Novartis, Qiagen, Pfizer, Roche, Targos MP Inc. RB is Co-Founder and Scientific Advisor for Targos Mol. Pathology Inc. RB is Testifying Advisor for MSD in GBA-Assessment for Pembrolizumab. RB has received funding from the Deutsche Krebshilfe for the Network Genomic Medicine. SMB has received speaker honoraria and personal fees from Pfizer, Novartis, Roche, Bayer, AstraZeneca, Molecular Health, GSK, MSD and Targos; speaker honoraria and non-financial support from BMS; non-financial support from Janssen. The authors declare no further conflict of interest. RC was supported by the Else Kröner-Fresenius Stiftung (2016-Kolleg.19). AMS, CA and BH were supported by Roche Pharma AG. The authors have no further conflict of interest to disclaim.
References
- Robinson KW, Sandler AB. The role of MET receptor tyrosine kinase in non-small cell lung cancer and clinical development of targeted anti-MET agents. Oncologist 18 (2013): 115-122.
- Armoiry X, Tsertsvadze A, Connock M, et al. Comparative efficacy and safety of licensed treatments for previously treated non-small cell lung cancer: A systematic review and network meta-analysis. PLoS One 13 (2018): e0199575.
- Mosele F, Remon J, Mateo J, et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: a report from the ESMO Precision Medicine Working Group. Ann Oncol (2020).
- Overbeck TR, Cron DA, Schmitz K, et al. Top-level MET gene copy number gain defines a subtype of poorly differentiated pulmonary adenocarcinomas with poor prognosis. Transl Lung Cancer Res 9 (2020): 603-616.
- Wolf J, Seto T, Han JY, et al. Capmatinib in MET Exon 14-Mutated or MET-Amplified Non-Small-Cell Lung Cancer. N Engl J Med 383 (2020): 944-957.
- Heydt C, Becher AK, Wagener-Ryczek S, et al. Comparison of in situ and extraction-based methods for the detection of MET amplifications in solid tumors. Comput Struct Biotechnol J 17 (2019): 1339-1347.
- Onozato R, Kosaka T, Kuwano H, et al. Activation of MET by gene amplification or by splice mutations deleting the juxtamembrane domain in primary resected lung cancers. J Thorac Oncol 4 (2009): 5-11.
- Shaw AT, Engelman JA. ALK in lung cancer: past, present, and future. J Clin Oncol 31 (2013): 1105-1111.
- Liu X, Jia Y, Stoopler MB, et al. Next-Generation Sequencing of Pulmonary Sarcomatoid Carcinoma Reveals High Frequency of Actionable MET Gene Mutations. J Clin Oncol 34 (2016): 794-802.
- Ou SH, Kwak EL, Siwak-Tapp C, et al. Activity of crizotinib (PF02341066), a dual mesenchymal-epithelial transition (MET) and anaplastic lymphoma kinase (ALK) inhibitor, in a non-small cell lung cancer patient with de novo MET amplification. J Thorac Oncol 6 (2011): 942-946.
- Frampton GM, Ali SM, Rosenzweig M, et al. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov 5 (2015): 850-859.
- Drilon A, Cappuzzo F, Ou SI, et al. Targeting MET in Lung Cancer: Will Expectations Finally Be MET? J Thorac Oncol 12 (2017): 15-26.
- Kawakami H, Okamoto I, Okamoto W, et al. Targeting MET Amplification as a New Oncogenic Driver. Cancers (Basel) 6 (2014): 1540-1552.
- Albertson DG, Collins C, McCormick F, et al. Chromosome aberrations in solid tumors. Nat Genet 34 (2003): 369-376.
- Hellman A, Zlotorynski E, Scherer SW, et al. A role for common fragile site induction in amplification of human oncogenes. Cancer Cell 1 (2002): 89-97.
- Camidge DR OG, Clark JW, Ou SI, et al. Crizotinib in patients (pts) with MET-amplified non-small cell lung cancer (NSCLC): Updated safety and efficacy findings from a phase 1 trial. J Clin Oncol 36 (2018): 9062.
- Schwab R, Petak I, Kollar M, et al. Major partial response to crizotinib, a dual MET/ALK inhibitor, in a squamous cell lung (SCC) carcinoma patient with de novo c-MET amplification in the absence of ALK rearrangement. Lung Cancer 83 (2014): 109-111.
- Lai GGY, Lim TH, Lim J, et al. Clonal MET Amplification as a Determinant of Tyrosine Kinase Inhibitor Resistance in Epidermal Growth Factor Receptor-Mutant Non-Small-Cell Lung Cancer. J Clin Oncol 37 (2019): 876-884.
- Dagogo-Jack I, Yoda S, Lennerz JK, et al. MET Alterations Are a Recurring and Actionable Resistance Mechanism in ALK-Positive Lung Cancer. Clin Cancer Res 26 (2020): 2535-2545.
- Shi R, Filho SNM, Li M, et al. BRAF V600E mutation and MET amplification as resistance pathways of the second-generation anaplastic lymphoma kinase (ALK) inhibitor alectinib in lung cancer. Lung Cancer 146 (2020): 78-85.
- Scagliotti G, Moro-Sibilot D, Kollmeier J, et al. A Randomized-Controlled Phase 2 Study of the MET Antibody Emibetuzumab in Combination with Erlotinib as First-Line Treatment for EGFR Mutation-Positive NSCLC Patients. J Thorac Oncol 15 (2020): 80-90.
- Kanemura H, Takeda M, Nakagawa K. Simultaneous targeting of MET overexpression in EGFR mutation-positive non-small cell lung cancer can increase the benefit of EGFR-TKI therapy? Transl Lung Cancer Res 9 (2020): 1617-1622.
- Schildhaus HU, Schultheis AM, Ruschoff J, et al. MET amplification status in therapy-naive adeno- and squamous cell carcinomas of the lung. Clin Cancer Res 21 (2015): 907-915.
- Cappuzzo F, Marchetti A, Skokan M, et al. Increased MET gene copy number negatively affects survival of surgically resected non-small-cell lung cancer patients. J Clin Oncol 27 (2009): 1667-1674.
- Go H, Jeon YK, Park HJ, et al. High MET gene copy number leads to shorter survival in patients with non-small cell lung cancer. J Thorac Oncol 5 (2010): 305-313.
- Paik PK, Felip E, Veillon R, et al. Tepotinib in Non-Small-Cell Lung Cancer with MET Exon 14 Skipping Mutations. N Engl J Med 383 (2020): 931-943.
- Castiglione R, Alidousty C, Holz B, et al. Comparison of the genomic background of MET-altered carcinomas of the lung: biological differences and analogies. Mod Pathol 32 (2019): 627-638.


 Impact Factor: * 4.1
Impact Factor: * 4.1 Acceptance Rate: 74.74%
Acceptance Rate: 74.74%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 