Fecal Microbial Profiling of Young Hirschsprung Disease Children After Pull-Through Operation
Kanokrat Thaiwatcharamas1, Watcharin Loilome2,3,4, Sinobol Chusilp1, Patchareeporn Tanming1, Poramate Klanrit2,3,4, Jutarop Phetcharaburanin2,3,4*
1Division of Pediatric Surgery, Department of Surgery, Khon Kaen University, Khon Kaen, Thailand
2Department of Biochemistry, Faculty of Medicine, Khon Kaen University, Khon Kaen, Thailand
3Cholangiocarcinoma Research Institute, Khon Kaen University, Khon Kaen, Thailand
4Khon Kaen University International Phenome Laboratory, Khon Kaen University, Thailand
*Corresponding Author: Jutarop Phetcharaburanin, Department of Biochemistry, Faculty of Medicine, Khon Kaen University, Khon Kaen, Thailand
Received: 02 September 2022; Accepted: 09 September 2022; Published: 26 October 2022
Article Information
Citation: Kanokrat Thaiwatcharamas, Watcharin Loilome, Sinobol Chusilp, Patchareeporn Tanming, Poramate Klanrit, Jutarop Phetcharaburanin. Fecal Microbial Profiling of Young Hirschsprung Disease Children After Pull-Through Operation. Journal of Surgery and Research 5 (2022): 568-577.
View / Download Pdf Share at FacebookAbstract
Introduction: This study aimed to characterize and compare the fecal microbial profiles between post pull-through Hirschsprung disease patients and healthy children aged younger than five years.
Method: Fresh fecal samples were collected from 10 post pull-through Hirschsprung disease patients and age range-matched 10 healthy children. Bacterial DNA obtained from fecal samples were analyzed using 16S rRNA Illumina MiSeq platform.
Results: Our findings demonstrated the significantly increased Firmicutes in Hirschsprung disease group compared to the healthy group (corrected p-value=0.007) at the phylum level. In addition, the Firmicutes/ Bacteroidetes ratio in Hirschsprung disease group was 4.8 times higher than that of its control counterpart. Bacilli were also significantly increased (corrected p-value=0.004), while Erysipelotrichi and Actinobacteria were significantly decreased in Hirschsprung disease patients (corrected p-value=0.04 and 0.03, respectively) at the class level. Moreover, functional analysis demonstrated that 20 enzymes and 18 Kyoto Encyclopedia of Genes and Genomes pathways were significantly different between groups (corrected p-value<0.01).
Conclusion: A distinct dysbiosis even when the aganglionic segment had already been removed was remarkably evident in young children with Hirschsprung disease, with a significant increase in Firmicutes and a nearly five-fold increase in proportion of Firmicutes/Bacteroidetes which may potentially be employed as the dysbiosis-related biological indicator.
Keywords
<p>Hirschsprung, Microbiome, Enterocolitis, Child</p>
Article Details
Introduction
The fundamental principle of Hirschsprung disease (HD) treatment is the removal of the aganglionic segment replacing with the ganglionic bowel while preserving the sphincter function [1]. However, after surgery, the patients still have a chance of Hirschsprung associated enterocolitis (HAEC) which ranges in severity from mild to life-threatening. The incidence of HAEC is approximately 17.3%- 35% [1-4]. The exact pathogenesis of HAEC remains unclear even though several causes have been proposed, including intestinal barrier dysfunction, abnormal innate immune response, and dysbiosis [5].
The microbiome refers to the full complement of microbiota, their genes, and genomes in a specific environment [6]. The human body harbors approximately 10-fold individual more microbes than our somatic and germ cells [7]. Most inhabit our gastrointestinal tract, which is estimated to contain 10 to 100 trillion microbial cells and over 1000 species [8]. The gut microbiota is a complex community of microbes. Most reside in the distal ileum and colon that possess the optimal niche with microbial nutrients (e.g., essential amino acids, vitamins) and indigestible compounds (e.g., plant polysaccharides) [9]. Although some microbes, bacteria, or viruses are reminiscent of pathogens, most intestinal microorganisms provide many benefits, including strengthening the gut barrier integrity, producing nutrients, promoting pathogen interception, and modulating host immunity [9,10]. A healthy intestinal environment is characterized by a diverse and abundant microbiota dominated by members of Bacteroidetes, Firmicutes, and Actinobacteria. In addition, other typical features include an intact mucosal barrier and high levels of short chain fatty acid (SCFA) production [11]. These balancing mechanisms can be disturbed as a result of an altered microbial composition and function or dysbiosis, often characterized by altering microbial community and its function, and disruption of mucus and epithelial barriers [12]. Dysbiosis is associated with many gastrointestinal disorders including HD [12-15].
In HD studies, the culture-dependent method has been used to show the association between HAEC and Clostridium difficile and rotavirus [16,17]. Unfortunately, only 1% of microbes can be cultured in laboratory conditions and this method cannot clarify the complexity of microbiomes and anaerobic bacteria. Therefore, many studies have employed other non-culture microbiome technologies, including polymerase chain reaction (PCR), amplified ribosomal DNA restriction analysis (ARDRA) and nucleotide sequencing to increase sensitivity and specificity [18-20]. Culture-independent studies in HD patients began around 2009 using a quantitative real-time PCR assay to quantify Bifidobacterial and Lactobacillus in feces of HD patients with or without HAEC and those of normal children [18]. Next-generation sequencing (NGS) or second-generation sequencing has been increasingly used to investigate microbial community in HD. The results demonstrated pathogenesis mechanisms and determined the relationships between the molecular biological patterns in HD and HAEC, suggesting that HAEC is more likely to be caused by dysbiosis rather than a single pathogen. Despite the fact that previous studies demonstrated the microbial differences between HD and normal children, results varied across different studies as microbial profiles can be significantly affected by age, birth route, geography, diet and drugs [13,21-23]. Therefore, we aimed to investigate the fecal microbial communities between post pull-through HD patients and normal children, with less than five years of age and similar ethnicity and geography.
Material and Methods
Patient recruitment
In this cross-sectional study, after providing information and receiving the written consent form, 10 Thai HD children (0-5 years old) who underwent the pull-through operation and followed up at Srinagarind Hospital, Khon Kaen University during October 2019 - September 2020 were enrolled. Ten Thai healthy children (0-5 years old) who were not diagnosed with gastrointestinal problems were included. All participants resided in the same geographical area. The diagnosis of HD was confirmed by histopathology. The location of the pulled-down ganglionic portion creating anastomosis was also confirmed by histopathology. Metadata collected in the current study included age, sex, weight, birth route, current diet, medical and surgical history, antibiotics, probiotic use, history of enterocolitis, and current problems. This study was approved by the Khon Kaen University Ethics Committee for Human Research (HE621496) and the written consent of the parents of all participants has been obtained and the blank consent form is in the supplemental file. Fresh stool was collected and stored frozen (−80 °C) until microbiome analysis.
Bacterial DNA extraction, amplicon preparation, and 16S sequencing
Bacterial DNA extraction was conducted using MoBio PowerSoil DNA isolation kit to isolate bacterial genomic DNA according to the manufacturer’s protocol. A total weight of approximately 250 mg was transferred to the PowerBead Tubes followed by the addition of 750 μL of bead solution and 60 μL of solution C1 containing sodium dodecylsulphate (SDS) prior to homogenization and cell lysis. The supernatant was transferred to a clean 2 ml collection tube and 250 μL of solution C2 was added as lysis buffer. After the mixture was incubated at 4°C for 5 minutes and centrifuged, 200 μL of solution C3 was added to terminate decomposition. Then the supernatant was transferred into a clean collecting tube and 1,200 μL of solution C4 added for binding the DNA to a spin filter. After centrifugation at 10,000 x g, 500 μL of washing solution C5 was added followed by centrifugation at 10,000 x g for 30 seconds. Then, 100 μL of the elution solution C6 was added to the center of the white filter membrane and centrifuged at room temperature for 30 seconds at 10,000 x g. For quantification of the DNA extracted from different samples, a spectrophotometer (Nanodrop) was used with 1.5% agarose gel electrophoresis for visualization. Amplification and sequencing of the V1-V2 region were conducted. In brief, 7.5 μL of genomic DNA from fecal samples were amplified using the primers 515F (5- GTGCCAGCMGCCGCGGTAA-3’) and 806R (5-GGACTACHVGGGTWTCTAAT-3’). The polymerase chain reaction (PCR) was performed using a thermocycler (T100TM Thermal Cycler, Bio-Rad) and Hotstar Master Mix (Qiagen, Germany). The PCR cycling conditions were as follows: initial denaturation at 95°C for 3 min; 25 cycles of denaturation at 95°C for 30 s; annealing at 55°C for 30 s; extension at 72°C for 30 s and the final extension step at 72°C for 5 min. The negative control (DNase free water) was applied in the DNA extraction and 16S amplification steps. Sequencing was performed on the Illumina MiSeq platoform (Macrogen, Korea), with read length of 301 base pair, paired-end. Following standard quality control and demultiplexing, the reads were processed using the QIIME2 TM version 2017.6.0 [24]. First, paired-end reads were joined, and size selected to reduce non-specific amplification. These reads were then grouped into operational taxonomic units (OTUs) based on sequence similarity using the Greengenes version 13.5 database and classified at ³ 97% identity of reads [25]. All relevant data can be found in Supporting information.
Statistical analysis
Microbiome analysis was performed using MicrobiomeAnalyst [26,27]. Data normalization was performed, and data scaling was using total sum scaling. The taxa abundance will be explored as relative abundance by stack bar chart. The differential abundance analysis was performed by edgeR algorithm with adjusted p-value cutoff equal or less than 0.05 for statistically significant. Clinical data analysis was conducted in STATA software version 14 using Fisher’s exact test for categorical data comparison [28]. The normal distribution of continuous data was analyzed using a t-test, while a Mann-Whitney U-test was performed for non-parametric data. Data are expressed as mean±SD for normal distribution continuous data. A p-value ≤ 0.05 was considered statistically significant. The predicted functional profiling of the 16S rRNA gene sequence was conducted using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) software and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database [29-32]. Differences in the abundance of bacterial genera or KEGG pathways between groups were analyzed using STAMP software [33]. A p-value ≤ 0.05 was considered statistically significant.
Results
Ten young HD children were followed up in our institute during the study period. There were seven (70%) and six males (60%) in the HD and in children that did not have HD groups, respectively. Median age was 2.39 years (1.11 to 3.7 years) in the HD group and 1.05 years (0.76 to 1.61 years) in the children that did not have HD group (Table 1). All children resided in the same geographical area. Five HD rectosigmoid type children underwent transanal endorectal pull-through as the definitive surgery, while the other three patients with long segment aganglionosis and two with total colonic aganglionosis underwent abdominal assisted in combination with endorectal pull-through. Five HD patients had recent antibiotics (within one week). Neither probiotics nor laxatives were administered in any patient. Six HD patients became ill with HAEC while collecting fecal samples (Table 2). For microbiome analysis, the microbial composition was assessed by comparing the relative abundances of taxa at phylum and class levels (Figure 1).
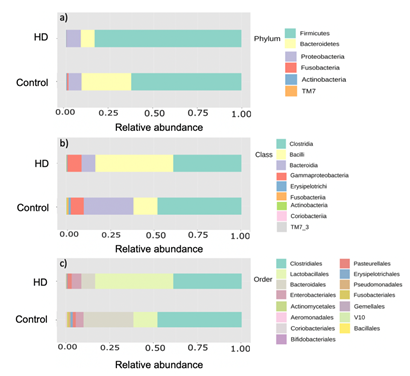
Figure 1: Taxonomic composition of fecal bacterial between Hirschsprung patients and normal control subjects. Stacked bar plot of taxonomic relative abundance at a) phylum b) class and c) order levels
The phylum level analysis showed that Firmicutes was the most prevalent in both the HD and non-HD groups (83.76% and 62.88%, respectively). The HD group had a reduction of Bacteroidetes, Actinobacteria, Fusobacteria and TM7, while Firmicutes and Proteobacteria were increased compared to the healthy group. Moreover, the Firmicutes/Bacteroidetes ratio (F/B ratio) in the HD group (10.66) was 4.8 times higher than that in the non-HD group (2.22). At the class level, Bacilli was the most prevalent in the HD group (44.46%), whereas fecal Clostridia predominated in the children that did not have HD group (47.89%). Differential abundance analysis showed that Firmicutes was significantly increased in the HD group compared to the children that did not have HD group (corrected p-value=0.007) at the phylum level, and Bacilli was also significantly increased at the class level (corrected p-value=0.004). Erysipelotrichi and Actinobacteria were significantly decreased in HD children (corrected p-value 0.02 and 0.03, respectively) compared to the healthy children (figure 2).
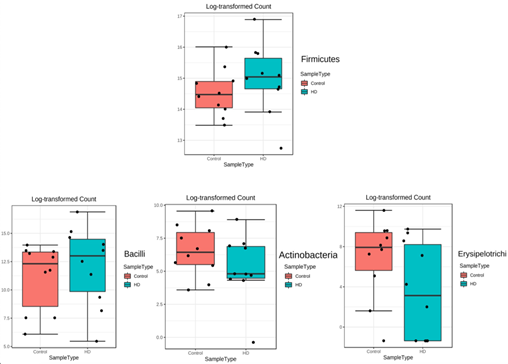
Figure 2: The box plots demonstrated the significantly different of fecal bacterial between Hirschsprung disease (HD) group and control group at the phylum and class level. A p-value less than 0.05 was considered as statistically significant.
Comparing between HD and HAEC group, the abundance at phylum level showed that Firmicutes was the most prevalent in both the HD with and without HAEC groups (85.47% and 82.77%, respectively). The HAEC group had a reduction of Bacteroidetes and increased Proteobacteria compared to the HD without HAEC patients (Figure 3). Moreover, the Firmicutes/Bacteroidetes ratio (F/B ratio) in the HAEC group was 1.89 times higher than in the HD group (11.22 vs. 5.95). At the class level, Bacilli was the most prevalent in the HAEC group (43.84%), whereas Clostridia was the most prevalent in the HD group (47.89%). Furthermore, the long segment aganglionosis group had a reduction of Bacteroidetes and increased of Proteobacteria compared to the rectosigmoid aganglionosis patients. At the class level, Clostridia was also the most prevalent in both rectosigmoid and long segment aganglionosis groups (64.37% and 33.39%, respectively) (Figure 3).
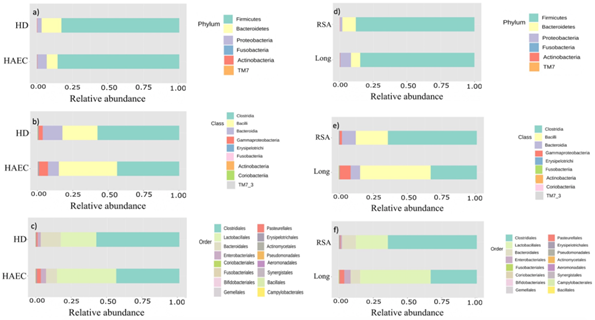
Figure 3: Taxonomic composition of fecal bacterial between Hirschsprung disease (HD) and Hirschsprung associated enterocolitis (HAEC) subjects, and between rectosigmoid (RSA) aganglionosis and long segment (Long) aganglionosis of HD patients. Stacked bar plot of taxonomic relative abundance at a) phylum b) class and c) order levels of fecal bacterial between HD and HAEC subjects. Stacked bar plot of taxonomic relative abundance at d) phylum e) class and f) order level of fecal bacterial between rectosigmoid (RSA) aganglionosis and long segment (Long) aganglionosis of HD patients.
In addition, differential abundance analysis between HD and HAEC group and between HD with rectosigmoid aganglionosis and long segment aganglionosis patients were also performed. There were no statistically significant differences in fecal bacterial analysis between HD and HAEC groups. Differential abundance analysis between each aganglionic segment showed that Bifidobacteriales and Pseudomonadales were significantly increased in the rectosigmoid aganglionosis HD patients compared to the long segment aganglionosis HD patients (corrected p-value=0.045) at the order level. In contrast, Actinomycetales was significantly decreased in rectosigmoid aganglionosis HD patients compared to the long segment aganglionosis HD patients (corrected p-value=0.045) (Figure 4). Functional analysis showed that the abundance of 38 KEGG endpoints corrected p-value=0.045 which included 20 enzymes and 18 KEGG pathways, was significantly different between groups (corrected p-value<0.01) (Figure 5).
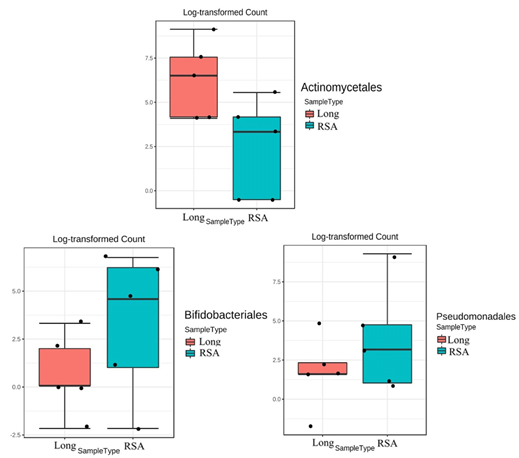
Figure 4: The box plots demonstrated the significantly different of fecal bacterial between rectosigmoid (RSA) and long segment aganglionosis (Long) in Hirschsprung disease patients at the order level. A p-value less than 0.05 was considered as statistically significant.
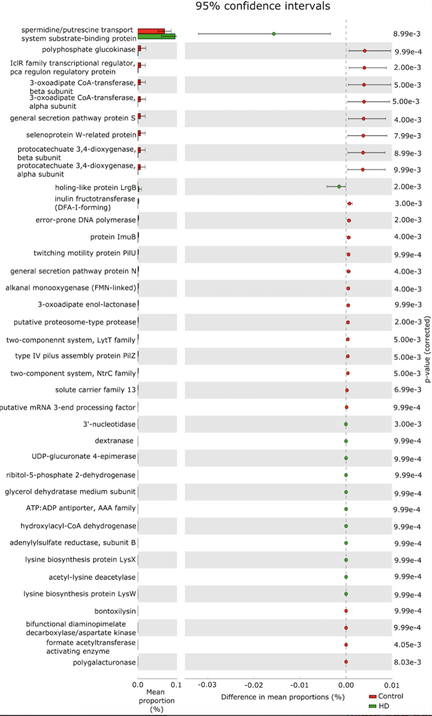
Figure 5: The relative abundance of functional pathways in the gut microbiota between control subjects and Hirschsprung disease (HD) patients. The Kyoto Encyclopedia of Genes and Genomes (KEGG) database functional categories are shown in the histograms. The corrected p-value determinations less than 0.01. Red and green denote the individual cases of control subjects and Hirschsprung disease patients.
Two pathways including spermidine/putrescine transport system substrate-binding protein (K11069) and holing-like protein LrgB (K05339), were significantly upregulated in HD patients than that of non-HD group (corrected p-value<0.01 with 95% CI did not include zero). Conversely, the KEGG pathway IclR family transcriptional regulator, pca regulon regulatory protein (K02624), general secretion pathway protein S (K02465), selenoprotein W-related protein (K07401), general secretion pathway protein N (K02463), protein ImuB (K14161), twitching motility protein PilU (K02670), and putative proteasome-type protease (K07395) were significantly downregulated in HD patients compared with non-HD groups (corrected p-value<0.01 with 95% CI did not include zero). Moreover, there were six enzymes, polyphosphate glucokinase (EC:2.7.1.63; K00886), 3-oxoadipate CoA-transferase, beta subunit (EC:2.8.3.6; K01032), 3-oxoadipate CoA-transferase, alpha subunit (EC:2.8.3.6; K01031), protocatechuate 3,4-dioxygenase, beta subunit (EC:1.13.11.3; K00449), protocatechuate 3,4-dioxygenase, alpha subunit (EC:1.13.11.3; K00448), inulin fructotransferase (DFA-I-forming) (EC:4.2.2.17; K10677), that were significantly predicted lower in HD group compared with non-HD group (corrected p-value<0.01 with 95% CI did not include zero).
Discussion
The gut microbiota is a complex community of microbes that are widely distributed in the gastrointestinal tract. Most microbes reside in the distal ileum and colon that contain the majority of microbial nutrients including essential amino acids and vitamins, together with indigestible componenets such as plant polysaccharides that can be microbially fermented into SCFAs [8]. An imbalance of the microbiota composition and its function, known as dysbiosis, could be involved with a number of gastrointestinal diseases, including HD [13-15,18]. After the advancement of analytical methods for microorganisms, HD-related microbiota has been studied although the results obtained from different studies varied [20,34-38]. Such different findings in the previous studies could result from the differences in age, geography, ethnicity and diet that are the important factors influencing gut microbiota [7,13]. In addition, the children’s microbial characteristics appear more diverse and converge towards the adult pattern after approximately 2.5 to 3 years of age, together with young post pull-through HD children being more susceptible to HAEC, leading to the age control of the sample collection in the current study [22,23]. In previous HD studies, the relative abundance of the phyla showed that Bacteroidetes predominated intraluminal content [20,35,37]. In contrast, the most predominant microbiota in both the HD and non-HD groups in our study was Firmicutes. This may be due to the differences in ethnicity, diet and the participant age range of our study compared with the others [20,34-38]. Even though Bacteroidetes was the second most abundant in the HD group, it was substantially decreased in the HD group compared with children that did not have HD. Having considered the F/B ratio in the HD group (10.66), it was 4.8 times higher than that in the non-HD group (2.22). Firmicutes and Bacteroidetes are the two major bacterial phyla in the gastrointestinal tract; thus, the ratio between these two phyla was formerly reported to associate with gut homeostasis and the alteration of this ratio also demonstrated to be associated with various diseases [39-41]. More interestingly, this proportion was reported to vary with age. In Ukrainian population, the median of F/B ratio in children group (0-9 years) was 0.69 and tended to increase with age compared to the elderly [42]. In addition, in Brazilian young children, the average F/B ratio was between 1.47 to 2.0, which was close to the ratio of the children that did not have HD group in our findings [43]. Therefore, to interpret microbial community results, our findings suggested that it may be necessary to consider the age of the study group, besides the disease itself. Comparison and classification of the taxa between the non-HD and HD groups showed that Firmicutes was significantly increased in the HD group compared to the non-HD group at the phylum level. This is probably because most of our young children with HD had history of HAEC that is consistent with a study by Li et al., comparing the intestinal microbiome content between HAEC and non-HAEC patients. They found that the relative abundance of Firmicutes was increased in HD with HAEC compared to patients without HAEC [37]. In addition, the comparison of the fecal microbial profiling between the lengths of the aganglionic segment had been demonstrated that there were significantly higher relative abundances of Bacteroidetes in rectosigmoid aganglionosis compared to total colonic aganglionosis whereas Proteobacteria was significantly higher in total colonic aganglionosis group [38]. Likewise, this study found the reduction of Bacteroidetes and increase of Proteobacteria in the long segment aganglionosis group compared to the rectosigmoid aganglionosis group. Our study found that Bacilli significantly increased in the HD group, while Erysipelotrichi and Actinobacteria significantly decreased. Although Bacilli are usually abundant in infants and are among the most common groups of probiotic bacteria, dysbiosis with a higher abundance of Bacilli was reported to be linked to diseases including HD [36,44,45]. Bacteria belonging to class Erysipelotrichi are in the phylum Firmicutes. Previous studies demonstrated the association of an increased abundance of this taxon with colon cancer, obesity, and choline deficiency-induced fatty liver disease [14,15,46]. In contrast, the study by Labbé and co-workers exhibited that in IBD patients, Erysipelotrichaceae, a family of the class Erysipelotrichi, significantly reduced compared to children that did not have HD [47]. Significantly increased relative abundances of Actinomyces, a genus of the class Actinobacteria, in HD patients compared to healthy children were evident in the previous study [36]. The decrease in Actinobacteria is probably due to antibiotics used for HAEC treatment in HD patients in the study [48]. The functional analysis depicted that the spermidine/putrescine transport system substrate-binding protein (K11069) were significantly upregulated in HD patients compared with the children that did not have HD. Spermidine and putrescine are the main polyamines in human cells that are mainly derived from ingested food and absorbed in the upper parts of the intestine, whereas polyamines found in the lower part of intestine are considered to be synthesized by the gut microbiota [49]. In general, the availability of polyamines in human cells contributes to tissue homeostasis of the gastrointestinal mucosa [50]. Hence, the dysregulation of polyamines, either under- or overexpression, can affect growth, aging and several diseases such as gastrointestinal cancer [50]. In bacteria, spermidine/putrescine transport system substrate-binding protein is in the ATP-binding cassette (ABC) transporters which include ABC importers for nutrient and micronutrient uptake, and ABC exporters for toxic substance excretion and drug resistance [30-32]. Holin-like protein LrgB appeared in hydrolases in a two-component system [51]. Hydrolysis activity regulates the growth rate by breaking apart the wall; thus, this activity plays an important role in many aspects of cell-wall growth, modification, and turnover in bacteria. It is, therefore, noteworthy to further investigate the host-microbial metabolic alteration in HD group. The limitations of this study included a small sample size and most HD patients, especially those who have a history of recurrent HAEC, require antibiotics for the treatment that may cause the microbial disturbance [36,38]. At present, from the current cross-sectional study, it is uneasy to determine whether the dysbiosis was altered by enterocolitis or by antibiotics. To tackle this challenge, further prospective study needs to be conducted.
Conclusion
Collectively, this study found significant differences in microbiota at both phylum and class levels between young Thai post pull-through HD and healthy children. There was a nearly five-fold increase in fecal F/B ratio of young post pull-through HD patients. Furthermore, the 38 KEGG endpoints of the functional analysis were found significantly altered between the two groups. Our findings revealed a distinct dysbiosis even when the aganglionic segment had already been removed in young HD children. Further longitudinal study with a larger sample size is still required to investigate the cause and effect of the gut microbiome in HD and HAEC patients.
Acknowledgments
The authors express gratitude to Professor Trevor N. Petney for editing the MS via the Publication Clinic KKU, Thailand. This work was supported by grants from Faculty of Medicine, Khon Kaen University (Grant No. IN63253) and Thailand Science Research and Innovation (TSRI) to KT and WL and by NSRF under the Basic Research Fund of Khon Kaen University to JP.
Financial interests
The authors declare they have no financial interests.
References
- Suita S, Taguchi T, Ieiri S, et al. Hirschsprung’s disease in Japan: analysis of 3852 patients based on a nationwide survey in 30 years. J Pediatr Surg 40 (2005): 197-202.
- Essam EBA, Coran AG, Blane CE, et al. Enterocolitis Associated With Hirschsprung’s Disease: A Clinical-Radiological Characterization Based on 168 Patients. J Pediatr Surg 30 (1995): 56-71.
- Menezes M, Puri P. Long-term outcome of patients with enterocolitis complicating Hirschsprung’s disease. Pediatr Surg Int 22 (2016): 316-318.
- Minford JL, Ram A, Turnock RR, et al. Comparison of Functional Outcomes of Duhamel and Transanal Endorectal Coloanal Anastomosis for Hirschsprung’s Disease. J Pediatr Surg 39 (2004): 161-165.
- Austin KM. The pathogenesis of Hirschsprung’s disease-associated enterocolitis. Semin Pediatr Surg 21 (2012): 319-327.
- Lederberg BJ, McCray AT. ‘Ome Sweet’ Omics- A Genealogical Treasury of Words. Sci 15 (2001): 8.
- Turnbaugh PJ, Ley RE, Hamady M, et al. The human microbiome project: exploring the microbial part of ourselves in a changing world. Nature 449 (2007): 804-810.
- Gill SR, Pop M, Deboy RT, et al. Metagenomic Analysis of the Human Distal Gut Microbiome. Science 312 (2006): 1355-1359.
- Srutkova D, Schwarzer M, Hudcovic T, et al. Bifidobacterium longum CCM 7952 promotes epithelial barrier function and prevents acute DSS-induced colitis in strictly strain-Specific manner. Plos One 10 (2015): 1-20.
- Jimenez AG, Ellermann M, Abbott W, et al. Diet-derived galacturonic acid regulates virulence and intestinal colonization in enterohaemorrhagic Escherichia coli and Citrobacter rodentium. Nat Microbiol 5 (2020): 368-378.
- Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464 (2010): 59-65.
- Chang JY, Antonopoulos DA, Kalra A, et al. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile - Associated Diarrhea. J Infect Dis 197 (2008): 435-438.
- O’Keefe SJD, Li J, Lahti L, et al. Fat, fibre and cancer risk in African Americans and rural Africans. Nat Commun 6 (2015): 198-205.
- Turnbaugh PJ, Ley RE, Mahowald MA, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444 (2006): 1027-1031.
- Chen W, Liu F, Ling Z, et al. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. Plos One 7 (2012): 90-97.
- Thomas DFM, Fernie DS, Bayston R, et al. Enterocolitis in Hirschsprung’s disease: A controlled study of the etiologic role of Clostridium difficile. J Pediatr Surg 21 (1986): 22-25.
- Wilson-Storey D, Scobie WG, McGenity KG. Microbiological studies of the enterocolitis of Hirschsprung’s disease. Arch Dis Child 65 (1990): 1338-1339.
- Shen DH, Shi CR, Chen JJ, et al. Detection of intestinal bifidobacteria and lactobacilli in patients with Hirschsprung’s disease associated enterocolitis. World J Pediatr 15 (2009): 56-62.
- De Filippo C, Pini-Prato A, Mattioli G, et al. Genomics approach to the analysis of bacterial communities dynamics in Hirschsprung’s disease-associated enterocolitis: A pilot study. Pediatr Surg Int 26 (2010): 465-471.
- Yan Z, Poroyko V, Gu S, et al. Characterization of the intestinal microbiome of Hirschsprung’s disease with and without enterocolitis. Biochem Biophys Res Commun 445 (2014): 269-274.
- Fallani M, Young D, Scott J, et al. Intestinal microbiota of 6-week-old infants across Europe: Geographic influence beyond delivery mode, breast-feeding, and antibiotics. J Pediatr Gastroenterol Nutr 51 (2010): 77-84.
- Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature 486 (2012): 222-227.
- Koenig JE, Spor A, Scalfone N, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci 108 (2011): 4578-4585.
- Bolyen E, Rideout JR, Dillon MR, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37 (2019): 852-857.
- DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a Chimera-Checked 16S rRNA Gene Database and Workbench Compatible with ARB. Appl Environ Microbiol 72 (2006): 5069-5072.
- Chong J, Liu P, Zhou G, et al. Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat Protoc 15 (2020): 799-821.
- Dhariwal A, Chong J, Habib S, et al. MicrobiomeAnalyst: A web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res 45 (2017): W180-W188.
- Stata Statistical Software: Release 14. College Station (2015).
- Langille MGI, Zaneveld J, Caporaso JG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31 (2013): 814-821.
- Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28 (2000): 27-30.
- Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci 28 (2019): 1947-1951.
- Kanehisa M, Furumichi M, Sato Y, et al. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res 49 (2021): D545-D551.
- Parks DH, Tyson GW, Hugenholtz P, et al. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30 (2014): 3123-3124.
- Tang W, Su Y, Yuan C, et al. Prospective study reveals a microbiome signature that predicts the occurrence of post-operative enterocolitis in Hirschsprung disease (HSCR) patients. Gut Microbes 23 (2020): 1-13.
- Frykman PK, Nordenskjöld A, Kawaguchi A, et al. Characterization of bacterial and fungal microbiome in children with Hirschsprung disease with and without a history of enterocolitis:A multicenter study. Plos One. 10 (2015): 3698-3712.
- Neuvonen MI, Korpela K, Kyrklund K, et al. Intestinal Microbiota in Hirschsprung Disease. J Pediatr Gastroenterol Nutr 67 (2018): 594-600.
- Li Y, Poroyko V, Yan Z, et al. Characterization of Intestinal Microbiomes of Hirschsprung’s Disease Patients with or without Enterocolitis Using Illumina-MiSeq High-Throughput Sequencing. Plos One 11 (2016): 4213-4229.
- Prato PA, Cavalieri D, Bartow-McKenney C, et al. A Metagenomics Study on Hirschsprung’s Disease Associated Enterocolitis: Biodiversity and Gut Microbial Homeostasis Depend on Resection Length and Patient’s Clinical History. Front Pediatr 1 (2019): 326.
- Duan M, Wang Y, Zhang Q, et al. Characteristics of gut microbiota in people with obesity. Plos One 16 (2021): e0255446.
- Wills ES, Jonkers DMAE, Savelkoul PH, et al. Fecal Microbial Composition of Ulcerative Colitis and Crohn’s Disease Patients in Remission and Subsequent Exacerbation. Plos One 9 (2014): e90981.
- Stojanov S, Berlec A, Štrukelj B. The influence of probiotics on the Firmicutes/Bacteroidetes ratio in the treatment of obesity and inflammatory bowel disease. Microorganisms 8 (2020): 1715.
- Vaiserman A, Romanenko M, Piven L, et al. Differences in the gut Firmicutes to Bacteroidetes ratio across age groups in healthy Ukrainian population. BMC Microbiol 20 (2020): 221-238.
- Chew C, Barros KV, Weffort VRS, et al. Gut Microbiota of Young Children Living in Four Brazilian Cities. Front Pediatr 8 (2020): 98-106.
- Marques TM, Wall R, Ross RP, et al. Programming infant gut microbiota: Influence of dietary and environmental factors. Curr Opin Biotechnol 21 (2010): 149-156.
- Lai HH, Chiu CH, Kong MS, et al. Probiotic Lactobacillus casei: Effective for managing childhood diarrhea by altering gut microbiota and attenuating fecal inflammatory markers. Nutrients 11 (2019): 1-15.
- Spencer MD, Hamp TJ, Reid RW, et al. Association between composition of the human gastrointestinal microbiome and development of fatty liver with choline deficiency. Gastroenterology 140 (2011): 976-986.
- Labbé A, Ganopolsky JG, Martoni CJ, et al. Bacterial bile metabolising gene abundance in Crohn’s, ulcerative colitis and type 2 diabetes metagenomes. Plos One 9 (2014): 69-78.
- Jakobsson HE, Jernberg C, Andersson AF, et al. Short-term antibiotic treatment has differing long- term impacts on the human throat and gut microbiome. Plos One 5 (2010): 1112-1118.
- Matsumoto M, Benno Y. The relationship between microbiota and polyamine concentration in the human intestine: A pilot study. Microbiol Immunol 51 (2007): 25-35.
- Timmons J. Polyamines and Gut Mucosal Homeostasis. J Gastrointest Dig Syst 3 (2013): 3698-3712.
- Brunskill EW, Bayles KW. Identification of LytSR-Regulated Genes from Staphylococcus Aureus. J Bacteriol 178 (1996): 709-716.


 Impact Factor: * 4.2
Impact Factor: * 4.2 Acceptance Rate: 72.62%
Acceptance Rate: 72.62%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 