Impaired Cellular and Antibody immunity after COVID-19 in Chronically Immunosuppressed Transplant Recipients
Chethan Ashokkumar1,2, Vinayak Rohan3, Alexander H Kroemer4, Sohail Rao5, George Mazariegos2, Brandon W Higgs2, Satish Nadig3, Jose Almeda5, Harmeet Dhani4, Khalid Khan4, Nada Yazigi4, Udeme Ekong4, Stuart Kaufman4, Monica M Betancourt-Garcia5, Kavitha Mukund6, Pradeep Sethi1, Shikhar Mehrotra3, Kyle Soltys2, Manasi S Singh3, Geoffrey Bond2, Ajai Khanna2, Mylarappa Ningappa2, Brianna Spishock1, Elizabeth Sindhi1, Neha Atale1, Maggie Saunders1, Prabhakar Baliga3, Thomas Fishbein4, Shankar Subramaniam6, and Rakesh Sindhi1,2*
1Plexision Inc., Pittsburgh, PA, USA
2Hillman Center for Pediatric Transplantation, University of Pittsburgh, PA, USA
3Medical University of South Carolina, Charleston, SC, USA
4Medstar Georgetown Transplant Institute, Washington, DC, USA
5DHR Health and DHR Health Institute for Research and Development, Edinburg, Tx, University of Houston, Houston, TX, USA
6University of California, San Diego, CA, USA
*Corresponding Author: Rakesh Sindhi, MD, FACS, UPMC Children’s Hospital of Pittsburgh, 4401 Penn Ave., FP-6/Transplant, Rm 6140 Pittsburgh, PA 15224, USA
Received: 20 September 2023; Accepted: 26 September 2023; Published: 18 October 2023
Article Information
Citation: Chethan Ashokkumar, Vinayak Rohan, Alexander H Kroemer, Sohail Rao, George Mazariegos, Brandon W Higgs, Satish Nadig, Jose Almeda, Harmeet Dhani, Khalid Khan, Nada Yazigi, Udeme Ekong, Stuart Kaufman, Monica M Betancourt-Garcia, Kavitha Mukund, Pradeep Sethi, Shikhar Mehrotra, Kyle Soltys, Manasi S Singh, Geoffrey Bond, Ajai Khanna, Mylarappa Ningappa, Brianna Spishock, Elizabeth Sindhi, Neha Atale, Maggie Saunders, Prabhakar Baliga, Thomas Fishbein, Shankar Subramaniam, and Rakesh Sindhi. Impaired Cellular and Antibody immunity after COVID-19 in chronically immunosuppressed transplant recipients. Journal of Surgery and Research. 6 (2023): 348-363.
View / Download Pdf Share at FacebookAbstract
Assessment of cellular immunity to the SARS-CoV-2 coronavirus is of great interest in chronically immunosuppressed transplant recipients (Tr), who are predisposed to infections and vaccination failures. We evaluated CD154-expressing T-cells induced by spike (S) antigenic peptides in 204 subjects-103 COVID-19 patients and 101 healthy unexposed subjects. Sreactive CD154+T-cell frequencies were a) higher in 42 healthy unexposed Tr who were sampled pre-pandemic, compared with healthy NT (p=0.02), b) lower in Tr COVID-19 patients compared with healthy Tr (p<0.0001) and were accompanied by lower S-reactive B-cell frequencies (p<0.05), c) lower in Tr with severe COVID-19 (p<0.0001), or COVID-19 requiring hospitalization (p<0.05), compared with healthy Tr. Among Tr with COVID-19, cytomegalovirus co-infection occurred in 34%; further, incidence of anti-receptor-binding-domain IgG (p=0.011) was lower compared with NT COVID-19 patients. Healthy unexposed Tr exhibit pre-existing T-cell immunity to SARS-CoV-2. COVID-19 impairs anti-S T-cell and antibody and predisposes to CMV co-infection in transplant recipients.
Keywords
<p>Cell-Mediated Immunity (CMI), SARS-CoV-2, Transplant recipients, Monocytic- and Polymorphonuclear-MDSC</p>
Article Details
Introduction
In chronically immunosuppressed transplant recipients (Tr), the status of immunity to SARS-CoV-2 is of great interest. This population is prone to life-threatening consequences of viral infection and failure of vaccination during periods marked by use of high- dose immunosuppression [1]. Lifelong use of anti-rejection immunosuppressants contributes to this impairment and may also limit post-infectious and post-vaccination immunity to SARS-CoV-2 [2,3]. Although antibodies can be demonstrated after natural SARS-CoV-2 infection and vaccination in the general population, this information is not as plentiful for Tr recipients [4-11]. Pre- existing T-cells that recognize SARS-CoV-2 are another component of immunity to this virus [12-15]. This type of immunity arises from prior exposure to human coronaviruses (hCoV), which account for 15% of seasonal flu and have structural similarities to SARS-CoV-2 [16,17]. Pre-existing cellular immunity may also compensate for impaired antibody responses to SARS-CoV-2 infection and vaccination, and aid in combating variant strains that are starting to emerge. T-cell immunity may also reassure those individuals wishing to re-engage with the general public, but who are unable to tolerate vaccination or fail to achieve a durable antibody response. Pre-existing cellular immunity to SARS-CoV-2 has been demonstrated in non-transplanted subjects (NT), but is not as well characterized in Tr recipients [12-15].
Recently proposed assays which measure T-cell immunity to SARS-CoV-2 may need to be modified to characterize T-cell immunity in Tr recipients. Some assays stimulate T-cells with those peptides representing the spike protein S, which have high affinity to well represented HLA specificities in a given population [13,14]. Such peptide mixtures can potentially overstimulate T-cells from individuals with these HLA specificities, but not T-cells from underrepresented individuals. Other assays also use the co-stimulators, anti-CD28 alone, or with anti-CD49d [12,14]. These adjunctive stimuli can also lead to an overestimate of T-cell immunity. Clinical decisions founded on such overestimates can be falsely reassuring in chronically immunosuppressed patients, and lead to errors in clinical judgement with adverse consequences. Some assays also use cytotoxic intracellular staining procedures, or only count those cells which co-express multiple markers as antigen-reactive. Because such “polyfunctional” T-cells are low frequency events, multi-marker assays require large numbers of cells from individuals with COVID-19, who can be severely lymphopenic. Another challenge is extrapolating findings from these early studies which use high affinity S antigenic peptides and costimulators, and most of which have been performed in NT subjects, who were convalescing or were not critically ill, to Tr recipients. These convalescent and non-critically ill immunocompetent subjects with COVID-19 demonstrated higher frequencies of S-reactive T-cells compared with those who were critically ill [12-15].
A clinically usable test design is exemplified by assays to measure T-cell response to cytomegalovirus (CMV). These assays use unselected peptide mixtures representing the entire antigenic sequence of interest, a single activation marker, and no costimulators [16-20]. Here, we use a minimal marker assay to characterize S-reactive T- and B-cells in healthy unexposed subjects and COVID-19 patients most of whom were hospitalized, with an emphasis on chronically immunosuppressed solid organ Tr recipients. A sizeable cohort of NT subjects is also included to enable robust conclusions and comparisons.
Methods
Human Subjects: COVID-19 patients were enrolled under IRB-approved protocols 2017-0365, Pro00101915, and 1551551 respectively, at three centers in Washington, DC, Charleston, SC, and Edinburg, TX, respectively. De-identified residual cryopreserved PBL samples were tested under IRB-exempt protocol, and samples from healthy-NT subjects were tested under IRB approved protocol 6774 in the reference laboratory (Plexision, Pittsburgh, PA). Healthy unexposed subjects, H-NT and H-Tr were tested using samples that were either obtained pre-pandemic, in 2019 or earlier, or were tested after confirming absence of symptoms suggestive of flu-like symptoms in the 6-month period prior to testing and a negative test for IgG to S and RBD antigens. COVID-19 patients, Tr or NT, were tested with samples obtained after confirmation of diagnosis with PCR.
Measuring SARS-CoV-2-reactive T-cell and B-cell subsets: All PBL samples were cultured alone (background), with 315 15-mer overlapping peptides with 11-mer overlap representing the 1273 amino acid spike antigen (test reaction), and with phorbol-myristic acid-Calcium ionophor (PMA, positive control) for 16 hours at 370C in 5% CO2 incubator. The peptide mixture consisted of two components mixed in equal parts-158 peptides representing the less conserved N-terminal sequence, S1, and 157 peptides representing the more conserved C-terminal sequence, S2, of the spike protein (JPT Peptides, Berlin, Germany). The S1 and S2 sequences respectively have 64% and 90% sequence homology with the SARS virus [21]. We used S-derived 1μg per stimulation condition for S1 and S2 respectively. For the S antigenic peptide mixtures, 1μg of S1 and 1μg of S2 were mixed to create the antigenic mixture. The culture medium contained fluorochrome-labeled antibody to CD154 (catalog #563886, BD Biosciences, San Jose, CA). Cells were acquired on the FACS-Canto II flow cytometer with blue, red and violet lasers after addition of fluorochrome labeled antibodies to CD3, CD4, CD8, and CD19 and the viability dye 7-aminoactinomycin-D (catalog #s 340662, 641407, 340692, 341103, 559925, respectively, BD Biosciences, San Jose, CA). The gating strategy is shown in figure S1. Scatterplots acquired from assay reaction conditions for CD3, CD4, CD8 and CD19 cells are shown in figure S1. Frequencies for each subset which were reactive to the S peptide mixture were analyzed further after subtracting corresponding background frequencies.
CMV- and mitogen-reactive T-cells: Previously described methods were used to measure frequencies of CMV- specific T-cells and mitogen-reactive T-cells that expressed CD154 in response to stimulation with the pp65-CMV antigenic peptides and PMA, respectively [18].
Serological assay to detect SARS-CoV-2 antibody: 96-well microtiter plates were coated overnight at 40C with commercially available S-protein (Cat # 46328, LakePharma, San Carlos, CA,) at 2 ug/ml, and blocked for 1hr with PBS-Tween + 3% milk powder (weight/volume). Precoated wells were incubated with diluted samples for 2 hours, followed by anti-human IgG (Fab specific) HRP labeled secondary antibody 1:3000 in PBS-T containing 1% milk for 1 hour. After adding substrate (OPD solution), followed by 50μl of 3M hydrochloric acid to stop the reaction, plates were read at 490 nm on a spectrophotometer. With all samples, inactivated human AB serum was used as a negative control, while monoclonal antibody CR3022 was used as a positive control. Results were read on a plate reader as optical density at 490 nm. An optical density of 0.45 or greater was considered a positive test as reported earlier [22].
Myeloid-derived suppressor cells (MDSC): MDSC represent early lineage cells that cause T-cell suppression and develop in response to lymphopenia and the inflammatory response to the viral infection [23-25]. Fluorochrome-labeled antibodies to the respective markers for each cell were used to characterize monocytic- and polymorphonuclear-MDSC (M-MDSC and P-MDSC). The respective phenotypes were CD14+HLADR- and CD15+CD14-CD11b+ [25]. Antibodies used were from Biolegend (Cat #307618,301906,301306, San Diego, CA) or BD Biosciences (Cat #563743, San Jose, CA).
Statistical methods: Descriptive statistics were used to summarize group features. Between group comparisons were performed with t-tests for unadjusted data and linear models to adjust for demographic variables.
Results
Human Subjects: Of 204 total subjects, 101 were healthy subjects, H-Tr or H-NT, and 103 had been recently diagnosed with COVID-19, Tr or NT. The 204 subjects included 74 Tr recipients, of whom 42 were sampled pre-pandemic in 2019 or earlier, and 32 had COVID-19. Of 130 NT subjects, 59 were H-NT subjects of whom twenty-five were sampled pre-pandemic and thirty-four were negative for COVID-19 by antibody testing. Seventy-one NT subjects had COVID-19. Compared with healthy unexposed subjects, COVID-19 patients were predominantly non-Caucasian (38/101 vs 83/103 non-Caucasians, p<0.001) males (47/101 vs 60/103 males, p=NS) and were significantly older (41 vs 54 years, p=7.7E-06). General demographics for all 204 subjects are summarized in table 1. Details including treatment and outcomes for COVID-19 patients are shown in table S1. The COVID-19 cohort was notable for 12 patients with mild disease (12%), and 33 (32%) with severe disease requiring mechanical ventilation. Among the severely ill, 23 or 70% died.
|
H-NT |
H-Tr |
NT |
Tr |
p value |
||||
|
N |
59 |
42 |
71 |
32 |
H-NT vs H-Tr |
NT vs Tr |
H-NT vs NT |
H-Tr vs Tr |
|
Age (Years) |
44 ± 2.1 |
43 ± 3.9 |
57 ± 2.0 |
51.1 ± 4.0 |
NS |
NS |
<0.05 |
NS |
|
Age range |
18 - 78 |
1.5 - 70.2 |
24 - 87 |
0.56 - 77 |
||||
|
Male: Female |
22:37 |
25:17 |
39:32 |
21:11 |
NS |
NS |
NS |
NS |
|
Race (C:AA:H:A) |
37:9:0:13 |
26:13:2:1 |
12:4:54:1 |
8:11:11:2 |
NS |
<0.05 |
<0.05 |
<0.05 |
|
Organ (L:K:LK) |
NA |
25:17:0 |
NA |
21:9:2 |
NA |
NA |
NA |
NS |
|
Alive:Dead |
59:0 |
42:0 |
53:18 |
27:5 |
NA |
NS |
NA |
NA |
|
Disease Severity (Intub:Hosp:Mild) Convalescent Plasma |
NA |
NA |
21:40:10 |
12:18:2 |
NA |
NS |
NA |
NA |
|
NA |
NA |
50 |
4 |
NA |
<0.05 |
NA |
NA |
|
|
Days from Dx |
8 ± 2 |
6.5 ± 2.3 |
NA |
NS |
NA |
NA |
||
|
Range (Days from Dx) |
0 to 94 days |
2 to 50 days |
||||||
Abbreviations: H-Tr: Healthy transplant, H-NT: healthy non-transplant, Tr-transplant recipients with COVID-19, NT-non-transplant patient with COVID-19, C: Caucasian, AA: African American, H: Hispanic, A: Asian, L: Liver transplant, K: Kidney transplant and LK: Liver-Kidney Transplant, Intub: Intubation, Hosp: Hospitalized, Mild: Mild, Dx: Diagnosis.
Table 1: General demographics of the study population.
S-reactive T-cells and B-cells co-express IFNγ and interleukin-6 (IL-6): We established that CD154 is co-expressed with IFNγ, a marker of cytotoxic T-cells, in PBL from 5 healthy human subjects, stimulated overnight with the S peptide mixture (Figure 1, Supplementary figure S2). Median (range) frequencies of S-reactive T-cells that co-expressed both markers were 3.1% (1.1-10.3), and greatly exceeded S-reactive T-cells that expressed either CD154, 0.2% (0.1-0.2) or IFNγ, 0% (0-0.1), respectively. Because nearly all S-reactive IFNγ+T-cells co-express CD154, using the single marker CD154 would overestimate S-reactive T-cells by 0.2% divided by the sum of 0.2% and 3.1% times 100, or 6%. Similarly, in four patients with COVID-19, S-reactive 196 CD154+IFNγ+T-cells were 0.74% (0.48-0.93), and greatly exceeded CD154+T-cells, 0.03% (0-197 0.08) or IFNγ+T-cells 0% (0-0) respectively (Figure 1). Because all S-reactive IFNγ+T-cells co-express CD154, using the single marker CD154 in patients with COVID-19 would overestimate S-reactive T-cells by 3.9%. In three healthy human subjects, median (range) frequencies of S-reactive B-cells that co-expressed IL-6 and CD154 were 4.9% (4.7-13.1) and greatly exceeded B-cells that expressed either CD154, 0.4% (0.3-0.4) or IL-6, 0% (0-0), respectively (Figure S3). Because all S-reactive IL-6+B-cells co-express CD154, using the single marker CD154 would overestimate S-antigen reactive B-cells by 7.54%. Therefore, S-reactive T- and B-cells capture all S-antigen reactive IFNγ+T-cells, and IL-6+B-cells, respectively, and overestimate these cell types by 3.9-7.5% and were used to test all samples [18,26]. The non-permeabilizing surface staining methods also preserve cell counts which can decrease by 40-50% with cell permeabilizing techniques required to detect intracellular cytokines.
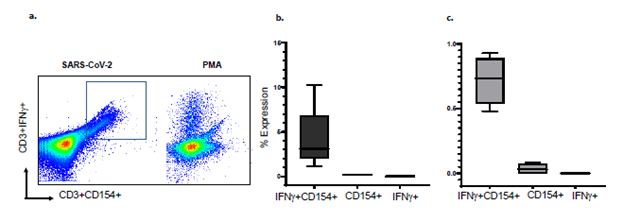
Figure 1: Flow cytometry scatterplots show a) expression of CD154 and IFNγ in S-reactive and PMA-reactive T-cells. Majority of IFNγ+ CD3 cells co-express CD154, Minimum and Maximum bar diagram are shown for b) healthy controls (n=5) and c). COVID-19 subjects (n=4). PMA=phorbol-myristic acid, a mitogen
Reproducibility: S-reactive CD154-expressing CD3, CD4, CD8 and CD19 cells were measured in duplicate assays performed on the same day, before and after 7 days of cryopreservation in liquid nitrogen, and before and after overnight storage or overnight shipment at ambient temperature. Mean coefficient of variation between duplicate assays was 2-10.6% in these various conditions (Tables S2-S5).
T- and B-cell responses to spike antigens are impaired with COVID-19 and increasing disease severity: Frequencies of S-reactive CD3, CD4, CD8 and CD19 B-cells were lower in 32 Tr recipients with COVID-19 compared with 42 H-Tr recipients (p<0.001) (Figure 2a-b, Table S6). S-reactive CD3 and S-reactive CD8 cell frequencies decreased progressively with increasing COVID-19 severity in Tr patients with COVID-19. This decrease achieved significance for hospitalized recipients and those with severe COVID-19, compared with healthy-Tr subjects (Figure 2c-d). S-reactive T-cell frequencies in Tr patients with mild COVID-19 were similar to those in healthy-Tr recipients (p=NS). NT patients with COVID-19 did not show statistically significant differences in the frequencies of the various S-reactive cells, compared with healthy-NT subjects. The sole exception consisted of lower S-reactive CD3 cells in NT patients with COVID-19, compared with healthy-NT subjects, p=0.045 (Figure 2e-f).
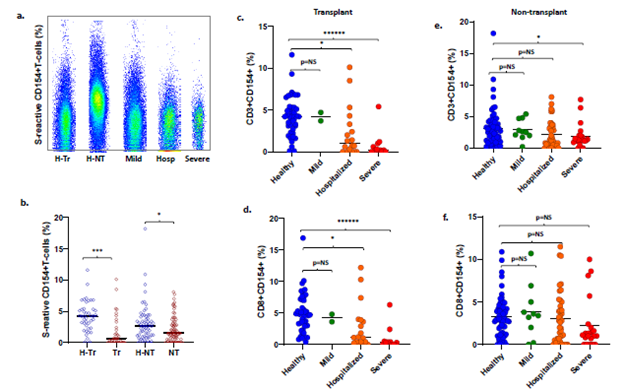
Figure 3: Optical density at 490 nm (OD490) for (a) Anti-RBD IgG and (b) Anti-spike IgG in transplant (Tr) and non-transplant (NT) patients with COVID-19. Dotted lines show the OD490 cutoff of 0.45 above which the tests are deemed positive. (c) S-reactive B-cell frequencies in healthy-NT, healthy-T and T and NT patients with COVID-19. (* represents p:value <0.05).
Conserved spike protein sequences have a larger contribution to SARS-CoV-2- specific T- and B-cell responses: Subsets of samples in from Tr and NT subjects were also stimulated with peptide mixtures representing the conserved C-terminal S2, and less conserved N-terminal S1 antigen. These subsets of samples were obtained from 63 of 74 Tr subjects and 104 out of 130 non-transplant (NT) subjects. In pairwise comparisons, frequencies of most S2-reactive CD154-expressing CD3, CD4, CD8 or CD19 cells were significantly higher in healthy-T and healthy-NT subjects compared with COVID-19 NT and COVID-19-T subjects (Table S7). Stimulation with the S1 peptide mixture elicited low frequency responses <1% or no responses in most samples. Despite this relative non-reactivity toward the S1 antigen, stimulation with the S antigen, which consisted of the S1 and S2 peptide mixtures elicited a larger response to stimulation than with either S1 or S2 alone (Figure S4).
Impaired antibody response to RBD in COVID-19 transplant patients: Of 74 COVID-19 patients with antibody measurements, 51 received convalescent plasma. IgG to spike antigen and RBD antigen were present in 49 of 51 (96%) and 47 of 51 (92%) patients, respectively. These subjects were excluded from analysis of humoral immunity. Among the remaining 23 patients who did not receive convalescent plasma, IgG to spike and RBD antigens were present in 21 (91%) and 16 (69.5%) patients, respectively. The incidence of anti-RBD IgG was significantly lower in transplant patients with COVID-19, 2 of 7 or 29%, compared with non- transplant patients, 14 of 16 or 88% (p=0.011) (Figure 3a). No differences were seen in the incidence of anti-spike IgG (5/7 or 71% vs 16/16 or 100%, p=NS) (Figure 3b). Subjects without and with anti-RBD antibody did not differ in timing of the sample from diagnosis (mean+/-SD 252 18+/-12.5 vs 12+/-12, p=0.258, NS, respectively), frequencies of S-reactive T-cells (mean 3.1+/-253 2.4% vs 1.8+/2%, p=0.225, NS, respectively), or proportions of patients requiring intubation (2/7 254 or 29% vs 4/16 or 25%, p=1.00, NS, respectively. S-reactive B-cell frequencies were also significantly lower in Tr and NT patients with COVID-19, compared with corresponding healthy subjects (Figure 3c).
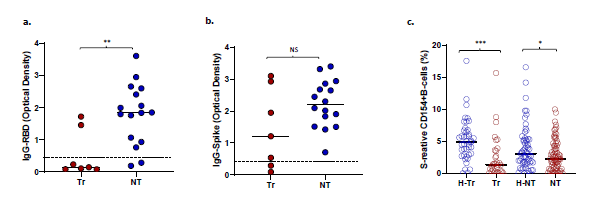
Figure 2: a. Flow cytometry scatterplots S-reactive CD3 cells in a representative healthy-transplant, healthy-non-transplant, Mild COVID-19, COVID-19 hospitalized (Hosp), and COVID-19 subject intubated for mechanical ventilation. b. Dot plots show frequencies of S-reactive T-cells (CD3) in healthy-transplant (H-Tr), COVID-19-transplant (Tr), healthy-non-transplant (H-NT) and COVID-19 non-transplant (NT) subjects. c-f. Dot plots show frequencies of S-reactive CD3 cells (c, e) and CD8 cells (d, f) in transplant (c, d) and non-transplant patients (e, f) with COVID-19 who have mild infection treated as outpatient, or are hospitalized or have severe infection. Corresponding frequencies from healthy transplant and non-transplant subjects are shown in each dot plot (* represents p:value <0.05).
Increased risk of CMV co-infection in transplant recipients: Of 32 Tr recipients with COVID-19, 11 (34%) experienced CMV infection-10 had CMV viremia and one had CMV hepatitis. To ascertain the basis of increased CMV risk, we measured frequencies of CMV-specific T-cells which express CD154 after stimulation with the pp65 antigenic peptide mixture in 61 subjects, as described previously [18]. CMV viremia is associated with decreased CMV-specific T-cell frequencies. Consistent with this known association, CMV-specific T-cell frequencies were significantly lower in 16 Tr recipients with COVID-19 compared with 13 healthy Tr recipients (0.5+/-0.4% vs 1.5+/-0.5%, p=3E-05, Figure 4a). CMV-specific T-cell frequencies were not significantly different between 6 NT subjects without and 26 NT subjects with COVID-19 (p=0.21, NS, Figure 4b). CMV infection did not occur in NT patients with COVID-19.
Increased circulating myeloid-derived suppressor cells (MDSC) during COVID-19: Twenty-four healthy and 29 COVID-19 patients were tested for circulating MDSCs. MDSC can suppress T-cells and are known to increase during viral infections. COVID-19 patients demonstrated higher frequencies of monocytic or M-MDSC (CD14+HLA-DR-) compared with healthy subjects (Median ± SEM, 39 ± 7.8% vs 2.95 ± 1.1%, p= 9.8 E-08) (Figure 4c). M-MDSC frequencies correlated negatively with S-reactive T-cell frequencies (Spearman’s r = -0.276, p= 0.045 Figure 4d). Polymorphonuclear or P-MDSC (CD15+CD14-CD11b+) frequencies were also higher in four COVID-19 subjects compared with 22 healthy subjects (median ± SEM, 64.2 ± 19.4% vs 1.25 ± 1.4%, p= 0.059, NS), and were negatively correlated with S-reactive T-cell frequencies (Spearman’s r = -0.518, p= 0.007).
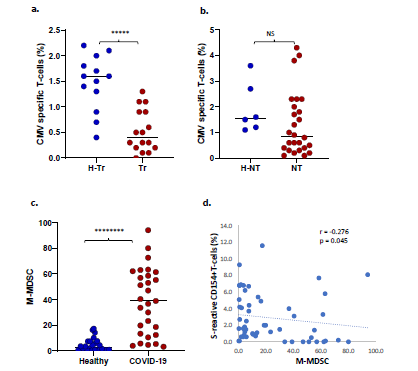
Figure 4: a. Dot plot shows CMV-specific T-cell frequencies among healthy transplant (H-Tr), healthy non-transplant (HNT), and COVID-19 patients with transplant (Tr) and without transplant (NT). b. frequencies of monocytic myeloid-derived suppressor cells (MDSC) in healthy unexposed subjects and COVID-19 patients. c. Correlation between frequencies of S-reactive CD154+T-cells and monocytic MDSC. (* represents p:value <0.05).
Discussion
Our study found that S-reactive T-cells are present in pre-pandemic PBL samples from chronically immunosuppressed transplanted (Tr) recipients. This type of pre-existing T-cell immunity has been reported previously in the general population and is also seen in our study population of healthy NT subjects [12-15]. Experimental evidence from previous studies implicates prior exposure to structurally similar human coronaviruses, which cause seasonal flu [16,17]. We speculate that this explanation also applies to our Tr recipient cohort. Unlike some previous studies, however, we observed lower S-reactive T-cell frequencies in COVID-19 patients compared with healthy unexposed individuals. This decrease was significant and most pronounced for Tr patients with COVID-19 compared with controls (Figure 2b-d). Further, compared with healthy-Tr subjects, Tr patients with COVID-19 also demonstrated a progressive decline in S-reactive T-cell frequencies with increasing disease severity, from hospitalization (p<0.05) to severe disease requiring intubation (p<0.0001). S-reactive T-cell frequencies in Tr patients with mild symptoms were in the same range as healthy unexposed Tr subjects (p=NS). S-reactive CD8 cells also demonstrated a similar disease-severity-dependent decline among Tr patients with COVID-19 (Figure 2d). Among NT patients with COVID-19, the decrease in S-reactive CD3 cell frequencies only achieved significance in those with severe COVID-19, compared with those without COVID-19 (Figure 2e).
Unlike previous studies, the majority of our COVID-19 patients, 91 of 103 or 88%, were hospitalized 58 without and 33 with severe disease requiring intubation for respiratory failure. Twenty three of 33 severely ill patients died. This distribution represents a more severely affected COVID-19 cohort and may explain lower mean T-cell frequencies in infected patients compared with those who were healthy. Loss of T-cell immunity to the virus has been observed in critically ill patients in some previous studies [12]. Previous reports have also shown higher S-reactive T-cell frequencies in convalescent patients compared with unexposed subjects [12-15]. These higher responses may be unique to the convalescent phase. Another reason for the higher T-cell responses in COVID-19 in some previous studies may be the use of peptides with high affinity for selected HLA specificities, with or without adjunctive co-stimulators. This approach may have elicited larger T-cell responses from memory subsets. Our study patients were sampled at an average interval of 12 days after diagnosis of COVID-19 and assayed using unselected peptide stimulators, without adjunctive co-stimulators.
In previous studies, S-reactive T-cell frequencies averaging <1% have been observed in healthy unexposed subjects, compared with roughly 3% in our studies [12-15]. Some of these studies counted S-reactive T-cells as those that co-expressed marker combinations like CD137 and CD69 but excluded S-reactive T-cells that expressed either marker alone, potentially underestimating viral antigen-specific T-cells [12,13]. For reasons stated in previous sections, we have modeled our assay on clinical assays which measure antiviral T-cell immunity by employing a single marker. S-reactive T-cell frequencies averaging 3% in our healthy unexposed subjects have also been observed among proliferating S-reactive T-cells in a previous study [14]. We cannot fully explain higher average frequencies in healthy unexposed Tr compared with NT subjects (mean 3.1 vs 4.2 %, p=0.042, Table S6). However, extended exvivo exposure of normal human PBL to pro-apoptotic anti-lymphocyte antibodies enriches apoptosis-resistant alloantigen-reactive CD154+T-cells among surviving PBL [27]. Thus, it is possible that exposure of T-cells to chronic immunosuppression may have contributed to an enrichment of S-reactive T-cells in PBL from Tr recipients.
The Tr recipient cohort with COVID-19 was noteworthy for CMV co-infection presenting as viremia in 10, and CMV hepatitis in one recipient for an incidence of 34%. CMV infection occurred at a median of 22 days (range 1-104 days) after diagnosis of COVID-19. Transplant recipients with COVID-19 also demonstrated lower frequencies of CMV-specific T-cells compared with NT COVID-19 patients, 0.4 ± 0.1 vs 0.85 ± 0.24, p=0.0048. Consistent with a lack of such differences in NT subjects, no CMV co-infections were reported in NT patients with COVID-19.
Of great interest is the observation that Tr recipients also demonstrated a lower incidence of IgG antibodies to the RBD component of the S protein after COVID-19 compared with NT recipients. These observations are consistent with impaired antibody response to COVID-19 vaccination in transplant patients [1]. The incidence of anti-S IgG antibodies was similar between the T and NT groups. The RBD sequence is a component of the less conserved N-terminal S1 sequence of the SARS-CoV-2 spike protein. The S1 protein has 60% sequence similarity to hCoV. As such, the RBD sequence may be less immunogenic when presented to the host immune system for the first time, compared with the more conserved C-terminal S2 sequence, which has 80% homology with hCoV. Test positivity was based on an OD490 of 0.45 or greater in the ELISA antibody binding assay. The amount of IgG antibody reflected by OD490 readings was also lower in Tr compared with NT patients for anti-spike IgG (p=0.16, NS) achieving significance for anti-RBD IgG (p<0.001) (Figure 3).
Suppressed cellular and antibody responses in Tr recipients may have other reasons. Recent studies have revealed increased circulating myeloid derived suppressor cells (MDSC), pyroptotic cell death and lymphopenia in COVID-19 patients [28-32]. MDSC are myeloid progenitors that expand in peripheral blood in response to lymphopenia and are known to suppress T-cells. Frequencies of monocytic and polymorphonuclear MDSC were higher in COVID-19 patients compared with healthy unexposed subjects. The associated decrease in S-reactive T-cells is reflected in significant negative correlations between S-reactive T-cells and MDSC.
It is noteworthy that T- and B-cell responses to the conserved S2 spike antigen more closely mirror responses to the entire spike protein in magnitude and predictive potential. Responses to the less conserved S1 protein were minimal or absent in healthy and COVID-19 patients. Possible reasons include a less immunogenic S1 sequence, or a slowly developing memory T-cell response to a SARS-CoV-2-specific antigen. Supportive evidence includes a lower incidence of IgG to RBD, a component of the S1 sequence, among chronically immunosuppressed transplant recipients. A possible explanation is that SARS-CoV-2 suppresses the antiviral T-cell response as evidenced by a simultaneous decrease in cellular immune response to CMV early after COVID-19. This suppression may occur via the induction of myeloid-derived suppressor cells which can suppress T-cells [29,30]. Longitudinal studies are needed to assess the relative role of cellular and humoral immunity to SARS-CoV-2 antigens at various intervals after natural infection and vaccination, especially among immunosuppressed patients.
In conclusion, transplant recipients demonstrate pre-existing T-cell immunity to SARS-CoV-2 as observed in the general population. Unique attributes of COVID-19 in transplant recipients include a) impaired T-cell immunity to SARS-CoV-2, to the greatest degree in those with increasing disease-severity, b) increased risk for CMV co-infection, and c) impaired antibody responses. Surveillance of CMV viral loads during COVID-19, and post-vaccination surveillance of antibody responses to confirm vaccine efficacy may be necessary in transplant recipients.
Acknowledgements:
NSF#2033307 (CA, Plexision) for assay standardization and reference range, NIH Grant number UL1 TR001450 (MUSC), intramural support from all participating institutions and Plexision.
Disclosure:
Cell based assays are based on University of Pittsburgh Patent 9606019, author: RS, which is licensed exclusively to Plexision, and in which University and RS own equity. CA and BH are paid consultants to Plexision. Other authors have nothing to disclose.
Data availability statement:
All data that underlie the results reported in this article (including study protocol) on individual participants will be made available to researchers who provide a methodologically sound proposal to the corresponding author.
Contributors and data management:
Study concept: RS and CA. Subject recruitment, interpretation of results, and editing and writing of manuscript: RV, AHK, JA, GM, SN, SR, HD, KK, MMBC, KS, GB, AK, MN, PB, TF HD, KK. Antibody testing: SN and SM. De-identification of subjects and summary of demographics: BS. CA performed and described CMI assays for SARS-CoV-2 antigens on de-identified samples. Data compilation, tabulation, cross-checks and summaries of flow cytometric cell counts and frequency for statistical analysis: BS, MS, NA and ES. Cytometry results and demographics were verified by PS, and transmitted to statistician BWH who merged the two datasets, performed all analyses and returned results and descriptions of analyses to PS and wrote and edited manuscript. KM and SS confirmed results of logistic regression with alternative linear models, edited and wrote manuscript. PS interpreted study results and relayed interpretations to RS for communication to all investigators. RS conceived the study, coordinated with centers and investigators, incorporated descriptions from other authors, wrote and edited manuscript with all authors.
Supplementary figures and tables
References
- Boyarsky BJ, Werbel WA, Avery RK, et al. Immunogenicity of a Single Dose of SARS-CoV-2 Messenger RNA Vaccine in Solid Organ Transplant Recipients. JAMA 325 (2021): 1784-1786.
- Akalin E, Azzi Y, Bartash R, et al. Covid-19 and Kidney Transplantation. N Engl J Med 382 (2020): 2475-2477.
- Choi M, Bachmann F, Naik MG, et al. Low Seroprevalence of SARS-CoV-2 Antibodies during Systematic Antibody Screening and Serum Responses in Patients after COVID-19 in a German Transplant Center. J Clin Med 9 (2020): 3401.
- Jackson LA, Anderson EJ, Rouphael NG, et al. An mRNA Vaccine against SARS-417 CoV-2 - Preliminary Report. N Engl J Med 383 (2020): 1920-1931.
- Mulligan MJ, Lyke KE, Kitchin N, et al. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature 586 (2020): 589-593.
- Folegatti PM, Ewer KJ, Aley PK, et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-426 blind, randomised controlled trial. Lancet 396 (2020): 467-478.
- Polack FP, Thomas SJ, Kitchin N, et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N Engl J Med 383 (2020): 2603-2615.
- Baden LR, El Sahly HM, Essink B, et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N Engl J Med 384 (2021): 403-416.
- Anderson EJ, Rouphael NG, Widge AT, et al. Safety and Immunogenicity of SARS-436 CoV-2 mRNA-1273 Vaccine in Older Adults. N Engl J Med 383 (2020): 2427-2438.
- Choe P, Kim K, Kang C, et al. Antibody Responses 8 Months after Asymptomatic or Mild SARS-CoV-2 Infection. Emerging Infectious Diseases 27 (2021): 928-931.
- Hartley GE, Edwards ESJ, Aui PM, et al. Rapid generation of durable B cell memory to SARS-CoV-2 spike and nucleocapsid proteins in COVID-19 and convalescence. Sci Immunol 5 (2020): eabf8891.
- Braun J, Loyal L, Frentsch M, et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature 587 (2020): 270-274.
- Grifoni A, Weiskopf D, Ramirez SI, et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 181 (2020): 1489-1501
- Sekine T, Perez-Potti A, Rivera-Ballesteros O, et al. Robust T Cell Immunity in Convalescent Individuals with Asymptomatic or Mild COVID-19. Cell 183 (2020): 158-168.
- Le Bert N, Tan AT, Kunasegaran K, et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 12 (2020): 235.
- Kahn JS, McIntosh K. History and recent advances in coronavirus discovery. Pediatr Infect Dis J 24 (2005): 223-227.
- Woldemeskel BA, Kwaa AK, Garliss CC, et al. Healthy donor T cell responses to common cold coronaviruses and SARS-CoV-2. J Clin Invest 130 (2020): 6631-6638.
- Ashokkumar C, Green M, Soltys K, et al. CD154-expressing CMV-specific T cells associate with freedom from DNAemia and may be protective in seronegative recipients after liver or intestine transplantation. Pediatr Transplant 24 (2020): e13601.
- Kumar D, Chin-Hong P, Kayler L, et al. A prospective multicenter observational study of cell-mediated immunity as a predictor for cytomegalovirus infection in kidney transplant recipients. Am J Transplant 19 (2019): 2505-2516.
- Rego K, Pereira K, MacDougall J, et al. Utility of the T-SPOT®.TB test’s borderline category to increase test resolution for results around the cut-off point. Tuberculosis 108 (2018): 178-185.
- Jaimes JA, Andre NM, Chappie JS, et al. Phylogenetic Analysis and Structural Modeling of SARS-CoV-2 Spike Protein Reveals an Evolutionary Distinct and Proteolytically Sensitive Activation Loop. J Mol Biol 432 (2002): 3309-3325.
- Xu G, Emanuel AJ, Nadig S, et al. Evaluation of Orthogonal Testing Algorithm for Detection of SARS-CoV-2 IgG Antibodies. Clin Chem 7 (2020): 563.
- O'Connor, Megan A et al. The Role of Myeloid-Derived Suppressor Cells in Viral Infection. Viral immunology 30 (2017): 82-97.
- Gabrilovich DI. Myeloid-Derived Suppressor Cells. Cancer Immunol Res 5 (2017): 3-8.
- Bronte V, Brandau S, Chen SH, et al. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat Commun 494 (2016): 12150.
- Ashokkumar C, Soltys K, Mazariegos G, et al. Predicting Cellular Rejection With a Cell-Based Assay: Preclinical Evaluation in Children. Transplantation 101 (2017): 131-140.
- Ashokkumar C, Sun Q, Ningappa M, et al. Antithymocyte globulin facilitates alloreactive T-cell apoptosis by means of caspase-3: potential implications for monitoring rejection-free outcomes. Transplantation 99 (2015): 164-1670.
- Schulte-Schrepping J, Reusch N. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell 182 (2020): 1419-1440.
- Agrati C, Sacchi A, Bordoni V, et al. Expansion of myeloid-derived suppressor cells in patients with severe coronavirus disease (COVID-19). Cell Death Differ 11 (2020): 1-12.
- Schulte-Schrepping J, Reusch N, Paclik D, et al. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell 182 (2020): 1419-1440.
- Feng S, Cheng X, Zhang L, et al. Myeloid-derived suppressor cells inhibit T cell activation through nitrating LCK in mouse cancers. Proc Natl Acad Sci 115 (2018): 10094-10099.
- Kroemer A, Khan K, Plassmeyer M, et al. Inflammasome activation and pyroptosis in lymphopenic liver patients with COVID-19. J Hepatol 73 (2020): 1258-1262.


 Impact Factor: * 4.2
Impact Factor: * 4.2 Acceptance Rate: 72.62%
Acceptance Rate: 72.62%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 