The Study on the Cellular Uptake Pathways of [Co(NTB)Cl]Cl-DNA Aggregates
Xiaoping Huang1*, Ang Li1, Lin Shao2
1College of Food Science and Technology, Wuhan Business University, Wuhan 430056, PR China
2Huazhong Agricultural University, Wuhan 430070, PR China
*Correspondingauthors: Xiaoping Huang, College of Food Science and Technology, Wuhan Business University, Wuhan 430056, PR China.
Received: 12 August 2025; Accepted: 21 August 2025; Published: 16 September 2025
Article Information
Citation: Xiaoping Huang, Ang Li, Lin Shao. The Study on the Cellular Uptake Pathways of [Co(NTB) Cl]Cl-DNA Aggregates. Journal of Biotechnology and Biomedicine. 8 (2025): 299-306.
DOI: 10.26502/jbb.2642-91280196
View / Download Pdf Share at FacebookAbstract
Metal complexes may become a new type of promising gene delivery systems because of their low cytotoxicity, structural diversity, controllable aqua- and lipo-solubility, and appropriate density and distribution of positive charges. We previously constructed a series of Metal (?) complexes of polybenzimidazoles, especially the dinuclear Co2+ complexes of polybenzimidazole ligands which were suggested to be the most potential nonviral gene carrier based on our previous work. The cellular uptake pathway of a gene vector is an important factor in transgene expression. Here in, we investigated the cellular uptake pathways of [Co(NTB)Cl]Cl-DNA aggregates into COS-7 cells by using the specific inhibitors. These inhibitors were applied to selectively inhibit uptake pathways by Clathrin-mediated endocytosis (CME), Caveolae-mediated endocytosis (CvME), macropinocytosis and microtubules. Meanwhile, we also investigated the cellular uptake pathways of PEI-DNA and lipofectamine 2000-DNA (Lipofect-DNA) aggregates into COS-7 cells as positive controls. Investigation of transgene expression showed that the [Co(NTB)Cl]Cl-DNA aggregates into COS-7 cells were mainly dependent on caveolae-mediated endocytosis (CvME) and macropinocytosis.
Keywords
<p>Metal complexes; [Co(NTB)Cl]Cl-DNA aggregates; Cellular uptake pathway</p>
Article Details
Introduction
Gene therapy is defined as the treatment of human disease by the transfer of genetic material into specific cells of the patients [1,2]. With the innovation of gene therapy and the development of clinical trials, the target of gene therapy has also gradually expanded from single-gene genetic diseases (albinism, hemophilia, etc.) to malignant tumors, infectious diseases, cardiovascular diseases, autoimmune diseases, metabolic diseases (diabetes, etc.) and other diseases [3]. There are two common kinds of gene vectors for gene therapy: viral gene vectors and non-viral gene vectors [4,5]. Compared with viral vectors, which has the potential of inducing immunogenicity, carcinogenicity and other safety issues, non-viral gene carriers are less immunotoxic, easier to be prepared and capable of carrying large amounts of genetic materials [6]. Non-viral gene delivery systems have been extensively studied over the last decades. Up to date, non-viral gene vectors mainly include cationic lipids, cationic polymers (such as polyethylenimine, PEI), peptides, cyclodextrins and their derivatives, nanoparticles carriers and metal complexes [7-11]. Among these vectors, metal complexes might stand out because of their low cytotoxicity, structural diversity, controllable aqua- and lipo-solubility, and appropriate density and distribution of positive charges [12,13]. Our group previously constructed a series of Metal (∥) complexes of polybenzimidazoles, such as the mononuclear or dinuclear Cu2+ and Co2+ and Ca2+ complexes of polybenzimidazole ligands, which had been confirmed to condense free DNA rapidly and the cellular uptake experiments successfully performed with the DNA condensates in many types of mammalian cells [14-22]. The cellular uptake pathway of a gene vector is an important factor in transgene expression [23]. Endocytosis is the process of transferring extracellular substances into cells through the deformation of plasma membrane, which includes phagocytosis and pinocytosis. It has been found that most non-viral gene vectors into cells are mainly dependent on pinocytosis. Pinocytosis includes clathrin-mediated endocytosis (CME), caveolae-mediated endocytosis (CvME), micropinocytosis and clathrin- and caveolin-independent endocytosis. In addition, some studies have shown that a few of non-viral gene vectors into cells are also dependent on microtubules and microfilaments [24-28].
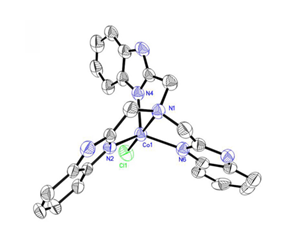
The commonly used method to study the uptake pathways of aggregates is using the specific inhibitors to inhibite endocytic pathway. Chlorpromazine can inhibit CME specifically by dissociating the clathrin lattice. Amiloride and derivatives inhibits macropinocytosis by inhibiting the Na+/H+ exchange protein. Cytochalasin D inhibits CvME by degrading the actin skeleton without affecting CME and Genistein is a tyrosine kinase inhibitor that also inhibits CvME. Nocodazole can degrade microtubules. The endocytic pathways of aggregates are confirmed by the expression level of luciferase in cells in the presence or absence of the specific inhibitors [29]. The cellular uptake pathways of different aggregates into different cells may be different. Lipofectamine2000 is a kind of polycationic carrier which can condense DNA effectively and has the higher transfection efficiency in many kinds of cells. Lipofectamine2000-DNA aggregates entered into HEK 293 cells via CME and macropinocytosis [30]. PEI2 complexes were taken up through CvME and macropinocytosis and PEI25 complexes were internalized mainly by CME, CvME and macropinocytosis in normal human foreskin fibroblast cells [31]. Meanwhile, the same aggregates entered into different cells via different cellular uptake pathways. For instance, PEI25 complexes were taken up through CvME and macropinocytosis in COS-7 cells [32]. However, the uptake pathways of the metal complexes which we constructed remains unclear. Based on our previous work, mononuclear or dinuclear Co2+ complexes of polybenzimidazole ligands included a variety of different complexes and the crystal structure of these complexes were obtained [33]. Among these complexes, [Co(NTB)Cl]Cl had a higher gene expression and a lower cytotoxicity when the ratio of [Co(NTB)Cl]Cl and DNA was 1 compared to other complexes (Fig.1) [20]. Thus, we investigated the cellular uptake pathways of [Co(NTB)Cl]Cl-DNA aggregates into COS-7 cells by applying the specific inhibitors of endocytic pathways and we found that the aggregates into COS-7 cells mainly relied on CvME and macropinocytosis.
Materials and methods
Materials
The pGL3 luciferase reporter gene vector (4818 bp, Promega) was purchased from WuHan TianYuanHuiDa biotechnology company. The pDNA was purified using an OMEGA Plasmid Mini Kit I (D6950-1). Enhanced BCA Protein Assay Kit (P0009), Methylthiazolyldiphenyl-tetrazolium bromide (MTT, ST1537) and Firefly Luciferase Reporter Gene Assay Kit (RG005) were obtained from Beyotime. Dimethyl sulfoxide (DMSO, 67-68-5), Chlorpromazine hydrochloride (69-09-0), Amiloride hydrochloride (1214-79-5), Genistein (446-72-0) and Nocodazole (31430-18-9) were purchased from Sigma-Aldrich. LipofectaminTM 2000 Transfection Reagent (Invitrogen™, 11668027) was purchased from Thermo Fisher Scientific. Poly (ethylenimine) (PEI, Ms 1800) was achieved from aladdin.
Cell line and cell cultures
African green monkey kidney fibroblast-like cell line COS-7 was obtained from WuHan Cell Collection Center. COS-7 cells were maintained in Dulbecco’s modified Eagle medium (Gibco) with 10% FBS (Gibco), 1% penicillin-streptomycin solution (Gibco). The cells were cultured under an atmosphere of 5% CO2/air at 370C.
Methods
Purification and Concentration of pGL3
Firstly, E.cole BL21 was cultured and pGL3 plasmid was activated. Then pGL3 plasmid was purified following the instructions of the OMEGA Plasmid Mini Kit. Finally, we tested absorbance of pGL3 at 260 nm and 280 nm by UV Spectrophotometer to calculate the purity and concentration of pGL3 according to the formula: C (DNA) = OD260 × dilution ratio × 50 μg/mL . Meanwhile, we also tested the configurations of pGL3 by agarose gel electrophoresis.
Preparation of the Aggregates
The PEI-DNA (1 μM of DNA, 3 μM of PEI) and [Co(NTB)Cl]Cl-DNA aggregates (1 μM of DNA, 1 μM of [Co(NTB)Cl]Cl ) were prepared for 30 min. Lipofect-DNA aggregates were prepared according to the manufacturer’s protocol.
Cytotoxicity assay
The working principle of MTT method is that the succinate dehydrogenase in the mitochondria of living cells can reduce the exogenous MTT to the purple crystalline formazan, which is insoluble in water but can be soluble in DMSO. There is an absorbance at 490 nm by the UV Spectrophotometer.
The cytotoxicity of inhibitors, inhibitors-complex and inhibitors-aggregates was evaluated by MTT assay. Briefly, the COS-7 cells were seeded at about 1000-10000 cells/well in 96-well plates, and incubated at 370C in a 5% CO2 humidified air atmosphere. The FBS-containing medium was replaced with a FBS-free medium when the cells grew to 90% confluence as a monolayer. The plates were divided into three groups: (1) Toxicity of four inhibitors: Chlorpromazine hydrochloride, Amiloride hydrochloride, Genistein and Nocodazole. (2) Toxicity of [Co(NTB)Cl]Cl complex in the presence of four inhibitors. (3) Toxicity of [Co(NTB)Cl]Cl-DNA, PEI-DNA and Lipofect-DNA aggregates in the presence of four inhibitors.
24 h later, MTT (5 mg/mL) was added to 96-well plates. After another 4 h of incubation, the MTT-containing medium was replaced by DMSO. The 96-well plates were oscillated for 10 min to fully dissolve the formazan crystal formed by living cells in the wells. The relative viability of the cells in each well was calculated by the absorbance of at 490 nm each well using the Biotek SynergyTM 2 Multi-detection Microplate Reader.
Cellular uptake pathways of aggregates assay
We used endocytic pathway inhibitors and the pGL3 luciferase reporter gene vector which strongly expressed luciferase in many types of mammalian cells to study the cellular uptake pathways of aggregates [33]. If the aggregates entered into cells was associated with some pathways, the luciferase expression of the samples was significantly reduced in the presence of the corresponding pathway inhibitor compared with the luciferase expression of the samples without pathway inhibitor.
COS-7 cells were seeded in 24-well plates at a density of 1×105 cells/well and incubated for overnight to reach 80% confluency. The FBS-containing medium was replaced with FBS-free medium when the cells grew to 80% confluence as a monolayer. Firstly, deterministic concentration of inhibitors was added to the corresponding plates: the concentration of Chlorpromazine hydrochloride was 2 mg/mL, Amiloride hydrochloride was 2.25 mg/mL, Genistein was 8 mg/mL and Nocodazole was 3 mg/mL. Put the 24-well plates into incubator and continue to cultivate for 1 h. Then, [Co(NTB)Cl]Cl-DNA aggregates, PEI-DNA aggregates and Lipofect-DNA aggregates were added to the corresponding plates. Put the 24-well plates into incubator and continued to cultivate for 24 h. Finally, the medium of 24-well plates was removed and the cells was washed with PBS. Adding the reporter gene lysis buffer to the plates and after lysis, cell debris was separated by centrifugation for 5 min at 5000 rpm and at 4 0C, supernatants were collected for fluorescence measurements. According to the manufacturer’s instructions of luciferase kid, the relative expression of intracellular luciferase was evaluated by the relative fluorescence intensity recorded by using the JASCO spectrofluorometer and a Biotek SynergyTM 2 Multi-detection Microplate Reader, respectively. The total fluorescence of intracellular proteins at 590 nm (excited at 338 nm) was digitally subtracted from each measurement of luciferase fluorescence. In fact, intracellular proteins do not emit any detectable fluorescence at this excitement wavelength.
Statistical analysis
All statistical tests were done using Microsoft Excel and origin 9, and the statistical analysis of data was indicated in each figure. For comparison between each sample group and its control group, unpaired Student’s t test was used. Parallel experiments were also indicated in each figure. Data were presented as means ± SD (indicated within each figure). P values less than 0.05 (*P < 0.05, **P < 0.01, ***P < 0.001) were considered statistically significant.
Results and Discussion
Purification and Concentration of pGL3
The concentration of DNA by measuring the absorbance of DNA at 260 nm. Meanwhile, the purity of DNA was determined by calculating OD260/OD280.
|
Serial Number |
260 nm |
280 nm |
OD260/OD280 |
CpGL3(μg/mL) |
|
1 |
0.7896 |
0.4202 |
1.879 |
789.6 |
|
2 |
0.6430 |
0.3419 |
1.880 |
643.0 |
|
3 |
0.6834 |
0.3628 |
1.884 |
683.4 |
|
4 |
0.7947 |
0.4218 |
1.884 |
794.7 |
Table 1: Results of the purity and concentration of pGL3 by UV Spectrophotometer.
We detected the concentration and purity of four pGL3 samples by UV Spectrophotometer. The results showed that OD260/OD280 ratio was between 1.8 and 1.9 indicating that the purity of pGL3 was suitable for the follow-up experiments and the concentration was between 550 and 800 μg/mL (Table1).
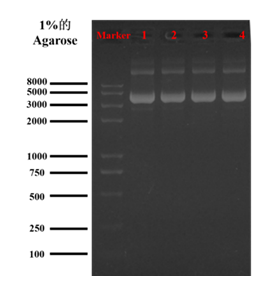
At a given electric field, the DNA migration rate depends on the DNA configuration and molecular weight. The migration rate of DNA with the same molecular weight in agarose gel electrophoresis is mainly related to the configuration of DNA. DNA configurations include covalently closed circle, linear form and open circular form. The migration rates of these three configurations in agarose gel were the fastest for covalently closed circle, followed by linear form and the slowest for open circular form [34]. The transient transfection efficiency was the highest under DNA covalently closed circle. Based on this theory, we used agarose gel electrophoresis to detect the forms of pGL3. The results of agarose gel electrophoresis showed that most of pGL3 retained their covalently closed circle configuration, which provided favorable conditions for the subsequent study of uptake pathways (Figure 2).
Cytotoxicity assay
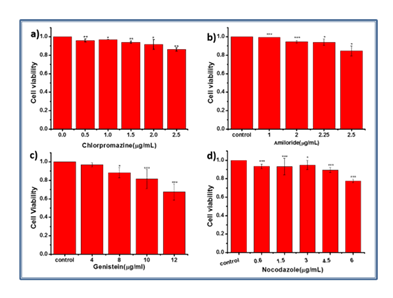
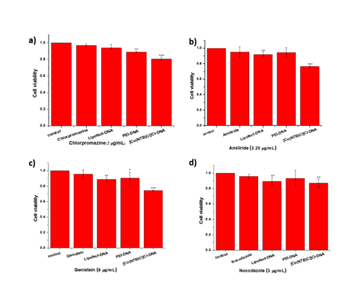
The cytotoxicity of specific inhibitors of endocytic pathway was evaluated by MTT assay with COS-7 cells under identical conditions. The multiple MTT tests showed that the cytotoxicity of inhibitors rose as a general trend with the inhibitor doses (Figure 3). Meanwhile, the cell viability was about 90% at a concentration of four inhibitors (Chlorpromazine hydrochloride was 2 mg/mL, Amiloride hydrochloride was 2.25 mg/mL, Genistein was 8 mg/mL, Nocodazole was 3 mg/mL) according to related references.
Figure 3: Cytotoxicity evaluation of four inhibitors. a) Chlorpromazine hydrochloride. The concentration was 0, 0.5, 1.0, 1.5, 2.0, 2.5 mg/mL. b) Amiloride hydrochloride. The concentration was 0, 1, 2, 2.25, 2.5 mg/mL. c) Genistein. The concentration was 0, 4, 8, 10, 12 mg/mL. d) Nocodazole. The concentration was 0, 0.5, 1.5, 3, 4.5, 6 mg/mL.(n≥3. *, P<0.05; **, P<0.01; ***, P<0.001)
Cytotoxicity was an important parameter to evaluate to ensure safe clinical trials with any gene carrier. Therefore, we evaluated the cytotoxicity of [Co(NTB)Cl]Cl complex in the presence of four inhibitors (Chlorpromazine hydrochloride was 2 mg/mL, Amiloride hydrochloride was 2.25 mg/mL, Genistein was 8 mg/mL, Nocodazole was 3 mg/mL) by MTT with COS-7 cells. The multiple MTT tests showed that the cytotoxicity of [Co(NTB)Cl]Cl complex rose with the concentration of complex doses. The results showed that the cell viability was above 80% when the concentration of complex was 1 mM (Figure 4).
Figure 4: Cytotoxicity evaluation of [Co(NTB)Cl]Cl complex in the presence of four inhibitors. a) Chlorpromazine hydrochloride was 2 mg/mL. b) Amiloride hydrochloride was 2.25 mg/mL. c) Genistein was 8 mg/mL. d) Nocodazole was 3 mg/mL. The concentration of [Co(NTB)Cl]Cl complex was 0, 0.5, 1, 1.5, 2, 2.5 mmol/L.(n≥3. *, P<0.05; **, P<0.01; ***, P<0.001)
We evaluated the cytotoxicity of [Co(NTB)Cl]Cl-DNA aggregates in the presence of four inhibitors (Chlorpromazine hydrochloride was 2 mg/mL, Amiloride hydrochloride was 2.25 mg/mL, Genistein was 8 mg/mL, Nocodazole was 3 mg/mL) by MTT with COS-7 cells. The multiple MTT tests showed that the cell viability was still above 75% (Figure 5). Therefore, the aggregates could be used for the following study of uptake pathways.
Figure 5: Cytotoxicity evaluation of PEI-DNA, Lipofect-DNA and [Co(NTB)Cl]Cl-DNA aggregates in the presence of four inhibitors. a) Chlorpromazine hydrochloride was 2 mg/mL, b) Amiloride hydrochloride was 2.25 mg/mL, c) Genistein was 8 mg/mL, and d) Nocodazole was 3 mg/mL. (n≥3. *, P<0.05; **, P<0.01; ***, P<0.001)
Cellular uptake pathways assay of [Co(NTB)Cl]Cl-DNA aggregates
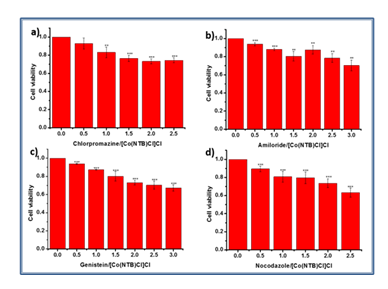
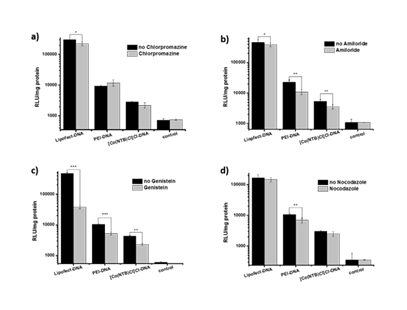
It has been widely reported that polyplexes enter mammalian cells through different endocytic pathways which used as a non-viral delivery system. We aimed to investigate the route of endocytic pathways of [Co(NTB)Cl]Cl-DNA. Meanwhile,we also explored the cellular uptake pathways of Lipofectamin and PEI. Endocytosis inhibitors such as Chlorpromazine, Amiloride, Genistein and Nocodazole were used to investigate the endocytic pathway of [Co(NTB)Cl]Cl-DNA, Lipofect-DNA and PEI-DNA.
COS-7 cells were incubated with polyplexes in the presence and absence of these inhibitors and the results were displayed in Figure 6. Compared to cells without inhibitors, treatment of COS-7 cells with Amiloride and Genistein reduced [Co(NTB)Cl]Cl-DNA mediated transfection at concentrations of 2.25 μg/mL and 8 μg/mL, respectively. On the other hand, the Chlorpromazine and Nocodazole did not affect the transfection efficiency of Cobalt-polybenzimidazole complex. The results showed that [Co(NTB)Cl]Cl-DNA aggregates were taken up through CvME and macropinocytosis in COS-7 cells. Chlorpromazine, Amiloride and Genistein reduced the transfection of Lipofect-DNA aggregates in cells compared to control. The transfection efficiencies of PEI-DNA aggregates in the presence of Amiloride, Genistein and Nocodazole were reduced obviously compared to cells without inhibitors. These results indicated that PEI-DNA aggregates were taken up into COS-7 cells mainly through CvME, macropinocytosis and microtubule. Lipofect-DNA aggregates were mainly relied on CvME, macropinocytosis and CME.
Discussion and Conclusions
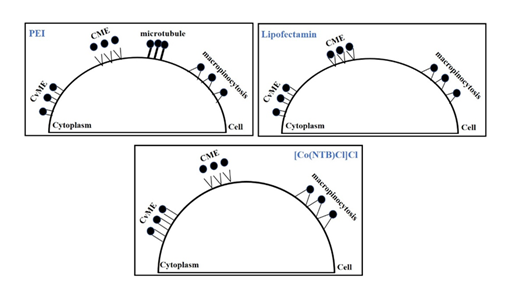
In summary, we had analyzed the cellular uptake pathways of the gene expression of [Co(NTB)Cl]Cl coated gene vector ([Co(NTB)Cl]Cl-DNA) using COS-7 cells. Meanwhile, we also analyzed the cellular uptake pathways of PEI and Lipofectamin 2000 into COS-7 cells (Figure 7).
Our study on the cellular uptake pathways of [Co(NTB)Cl]Cl complexes lay a foundation for the future studies on the uptake pathways of other vectors in different cells. They made us to better understand the mechanism of the non-viral gene vectors entering into cells. Meanwhile, this study could provide certain information support for improving the uptake rate of existing gene vectors and designing more non-viral gene vectors. It was important for the study to design more gene delivery systems. Therefore, this work would help to design more new non-viral gene carriers based on simple inorganic complexes.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was financially supported by Scientific Research Program of the Hubei Provincial Department of Education (B2024278) and Doctoral Foundation program of Wuhan Business University (X2023051508562045).
References
- Qiu K, Yu B, Huang H, et al. Tetranuclear ruthenium(II) complexes with oligo-oxyethylene linkers as one- and two-photon luminescent tracking non-viral gene vectors. Dalton Trans 44 (2015): 7058-65.
- Shahryari A, Saghaeian Jazi M, Mohammadi S, et al. Development and Clinical Translation of Approved Gene Therapy Products for Genetic Disorders. J Front Genet 10 (2019):
- Gonçalves GAR, Paiva RMA. Gene therapy: advances, challenges and perspectives. Einstein (Sao Paulo). 15 (2017): 369-375.
- Athanasopoulos T, Munye MM, Yáñez-Muñoz RJ. Nonintegrating Gene Therapy Vectors. Hematol Oncol Clin North Am 31 (2017): 753-770.
- Sung YK, Kim SW. Recent advances in the development of gene delivery systems. Biomater Res 23 (2019):
- Hooshmand SE, Jahanpeimay Sabet M, Hasanzadeh A, et al. Histidine-enhanced gene delivery systems: The state of the art. J Gene Med 24 (2022):
- Chen M, Tang Y, Wang T, et al. Enhanced gene delivery of low molecular weight PEI by flower-like ZnO microparticles. Mater Sci Eng C Mater Biol Appl 69 (2016): 1367-1372.
- Zu H, Gao D. Non-viral Vectors in Gene Therapy: Recent Development, Challenges, and Prospects. AAPS J 23 (2021):
- Gallops C, Ziebarth J, Wang A polymer physics perspective on why PEI is an effective nonviral gene delivery vector. Polymers in therapeutic delivery, United States (2020): 1–12.
- Sevimli S, Sagnella S, Kavallaris M, et al. Assessment of cholesterol-derived ionic copolymers as potential vectors for gene delivery. Biomacromolecules 14 (2013): 4135-4149.
- Dai Y, Xiao H, Liu J, et al. In vivo multimodality imaging and cancer therapy by near-infrared light-triggered trans-platinum pro-drug-conjugated upconverison nanoparticles. J Am Chem Soc 135 (2013): 18920-9.
- Bhat SS, Revankar VK, Khan A, et al. Luminescent Ruthenium(II) Polypyridyl Complexes as Nonviral Carriers for DNA Delivery. Chem Asian J 12 (2017): 254-264.
- Li C, Tian H, Duan S, et al. Controllable DNA condensation-release induced by simple azaheterocyclic-based metal complexes. J Phys Chem B 115 (2011): 13350-13354.
- Meng X, Liu L, Zhou C, et al. Dinuclear copper(II) complexes of a polybenzimidazole ligand: their structures and inductive roles in DNA condensation. Inorg Chem 47 (2008): 6572-6574.
- Liu L, Zhang H, Meng X, et al. Dinuclear metal(II) complexes of polybenzimidazole ligands as carriers for DNA delivery. Biomaterials 31 (2010): 380-391.
- Liu C, Yu S, Li D, et al. DNA hydrolytic cleavage by the diiron(III) complex Fe(2)(DTPB)(mu-O)(mu-Ac)Cl(BF(4))(2): comparison with other binuclear transition metal complexes. Inorg Chem 41 (2002): 13-22.
- Liu CL, Wang M, Zhang TL, et al. DNA hydrolysis promoted by di- and multi-nuclear metal complexes. Coord Chem Rev 248 (2004): 147-168.
- Liu CL, Wang DNA hydrolytic cleavage catalyzed by synthetic multinuclear metallonucleases. Dalton Trans 38 (2009): 227-239.
- Meng XG, Liu L, Zhang H, et al. Tris(benzimidazolyl)amine-Cu(II) coordination units bridged by carboxylates: structures and DNA-condensing property. Dalton Trans 40 (2011): 12846-12855.
- Jiang R, Yin J, Hu S, et al. Cobalt(II)-polybenzimidazole complexes as a nonviral gene carrier: effects of charges and benzimidazolyl groups. Curr Drug Deliv 10 (2013): 122-133.
- Huang X, Dong X, Li X, et al. Metal-polybenzimidazole complexes as a nonviral gene carrier: effects of the DNA affinity on gene delivery. J. Inorg. 129 (2013): 102-111.
- Yin J, Meng X, Zhang S, et al. The effect of a nuclear localization sequence on transfection efficacy of genes delivered by cobalt(II)-polybenzimidazole complexes. 33 (2012): 7884-7894.
- Yamada Y, Hashida M, Harashima H. Hyaluronic acid controls the uptake pathway and intracellular trafficking of an octaarginine-modified gene vector in CD44 positive- and CD44 negative-cells. Biomaterials 52 (2015): 189-98.
- Miernikiewicz P, Dąbrowska K. Endocytosis of Bacteriophages. Curr Opin Virol 52 (2022): 229-235.
- Gonçalves C, Mennesson E, Fuchs R, et al. Macropinocytosis of polyplexes and recycling of plasmid via the clathrin-dependent pathway impair the transfection efficiency of human hepatocarcinoma cells. Mol Ther 10 (2004): 373-85.
- Khalil IA, Kogure K, Akita H, et al. Uptake pathways and subsequent intracellular trafficking in nonviral gene delivery. Pharmacol Rev 58 (2006): 32-45.
- Doherty GJ, McMahon Mechanisms of endocytosis. Annu Rev Biochem 78 (2009): 857-902.
- Icard P, Lincet H. A global view of the biochemical pathways involved in the regulation of the metabolism of cancer cells. Biochim Biophys Acta 1826 (2012): 423-433.
- Peterson JR, Mitchison TJ. Small molecules, big impact: a history of chemical inhibitors and the cytoskeleton. Chem Biol 9 (2002): 1275-1285.
- Hu Q, Wang J, Shen J, et al. Intracellular pathways and nuclear localization signal peptide-mediated gene transfection by cationic polymeric nanovectors. 33 (2012): 1135-45.
- Hsu CY, Uludağ H. Cellular uptake pathways of lipid-modified cationic polymers in gene delivery to primary cells. Biomaterials 33 (2012): 7834-7848.
- van der Aa MA, Huth US, Häfele SY, et al. Cellular uptake of cationic polymer-DNA complexes via caveolae plays a pivotal role in gene transfection in COS-7 cells. Pharm Res 24 (2007): 1590-1598.
- Stanić V, Arntz Y, Richard D, et al. Filamentous condensation of DNA induced by pegylated poly-L-lysine and transfection efficiency. Biomacromolecules 9 (2008): 2048-2055.
- Lee PY, Costumbrado J, Hsu CY, et al. Agarose gel electrophoresis for the separation of DNA fragments. J Vis Exp 62 (2012):


 Impact Factor: * 5.3
Impact Factor: * 5.3 Acceptance Rate: 75.63%
Acceptance Rate: 75.63%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 