Non-enzymatic Methylation of Cytosine in RNA by S-adenosylmethionine and Implications for the Evolution of Translation
Bruce K Kowiatek*
Blue Ridge Community and Technical College, Martinsburg, WV, USA
*Corresponding Author: Bruce K. Kowiatek, Blue Ridge Community and Technical College, 13650 Apple Harvest Dr, Martinsburg, WV, USA
Received: 14 June 2019; Accepted: 01 July 2019; Published: 23 July 2019
Article Information
Citation:
Bruce K Kowiatek. Non-enzymatic Methylation of Cytosine in RNA by S-adenosylmethionine and Implications for the Evolution of Translation. Journal of Biotechnology and Biomedicine 2 (2019): 048-056.
DOI: 10.26502/jbb.2642-9128009
View / Download Pdf Share at FacebookAbstract
The non-enzymatic methylation of cytosine (C) to form 5-methylcytosine (5-mC) in deoxyribonucleic acid (DNA) by the intracellular methyl group donor S-adenosylmethionine (SAM), resulting in S-adenosylhomocysteine (SAH) and minor thymine (T) via spontaneous deamination, implicated in certain point mutagenic cancers, has been widely known since the 1980s, as has the proposed Watson-Crick mechanism of the adenine (A) moiety of SAM base-pairing with T or uracil (U). Such analogous base-pairing and non-enzymatic methylation in ribonucleic acid (RNA), however, has not been as widely addressed, particularly with respect to the origins and evolution of the process of translation initiation in the context of the hypothesized RNA world that preceded the current DNA-protein world. It is posited here, with spectrophotometric evidence put forth, that such base-pairing and non-enzymatic methylation with subsequent deamination in RNA may constitute a rudimentary form of metabolism and self-replication with implications for the origins and evolution of translation initiation, possibly including the origin and evolution of the transfer RNA (tRNA) molecule.
Keywords
<p>S-adenosylmethionine, Non-enzymatic methylation, DNA, RNA, Evolution</p>
Article Details
Abbreviations:
DNA-deoxyribonucleic acid; SAM-S-adenosylmethionine; SAH-S-adenosyl-homocysteine; RNA-ribonucleic acid; tRNA-transfer RNA
1. Introduction
The non-enzymatic methylation of cytosine (C) to form 5-methylcytosine (5-mC) in deoxyribonucleic acid (DNA) by the intracellular methyl group donor S-adenosylmethionine (SAM) (Figures 1 and 4), resulting in S-adenosylhomocysteine (SAH) and minor thymine (T) via spontaneous deamination (Figure 2), implicated in certain point mutagenic cancers, has been widely known since the 1980s [1], as has the proposed Watson-Crick mechanism of the adenine moiety (A) (Figure 3) of SAM base-pairing with T or uracil (U) (Figure 5) [2]. Such analogous base-pairing and non-enzymatic methylation in ribonucleic acid (RNA), however, has not been as widely addressed, particularly with respect to the origins and evolution of the process of translation initiation in the context of the hypothesized RNA world that preceded the current DNA-protein world [3]. It is posited here, with spectrophotometric evidence put forth, that such base-pairing and non-enzymatic methylation with subsequent deamination in RNA may constitute a rudimentary form of metabolism and self-replication with implications for the origins and evolution of translation initiation, possibly including the origin and evolution of the transfer RNA (tRNA) molecule (Figure 6).
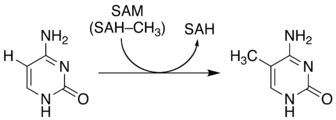
Figure 1: Non-enzymatic methylation of C via SAM to form 5-mC and SAH.
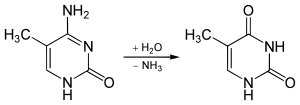
Figure 2: Spontaneous deamination of 5-mC to form T.
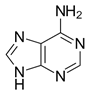
Figure 3: Adenine moiety (A) of SAM.
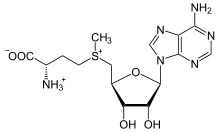
Figure 4: S-adenosylmethionine (SAM).
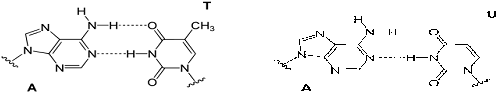
Figure 5: Watson-Crick base-pairing mechanism of the adenine moiety (A) of SAM with T and U.
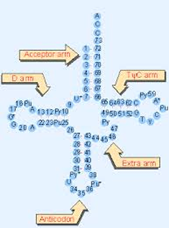
Figure 6: Typical tRNA molecule with typically conserved bases. Note conserved TΨC bases in the T-loop section of the TΨC arm. Abbreviations: Ψ=pseudouridine, Py=pyrimidine (C or U), and Pu=purine (A or G).
During translation initiation, the small ribosomal subunit binds to the messenger RNA (mRNA) initiation sequence, whereupon a tRNA molecule, universally carrying the amino acid component of SAM, methionine (Met, M), formylated to N-formylmethionine (fMet) in bacteria (Figure 7), and binds to the mRNA triplet nucleotide (nt) Adenine Uracil Guanine (AUG) initiation codon sequence (Figure 8); while other mRNA initiation codon sequences exist in prokaryotes such as archaea and bacteria, all still contain UG as their second and third respectively, and all still carry Met or fMet, regardless of their position in the genetic code [4].
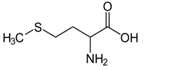
Figure 7: Methionine (Met, M), the amino acid component of SAM; N-formylmethionine (fMet) found in bacteria.
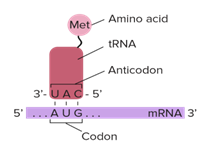
Figure 8: Translation initiation taking place on the small ribosomal subunit.
2. Experimental
All research was conducted at Blue Ridge Community and Technical College in Martinsburg, WV, USA from January of 2019 through May of 2019. All pH measurements were made using a Horiba TwinpH waterproof B-213 Compact pH meter. All spectrophotometric data was obtained using a Beckman Coulter DU 530 UV/Vis Spectrophotometer. UV spectra were compared to the standard peak Absorbance (logarithm epsilon) value of 267 nanometers (nm) for T [5]. All Pyrex glassware was sterilized at 130° C for one hour [6] via autoclave using a Quincy Lab Inc. Model 30 GC Lab Oven. All chemical supplies were purchased from Millipore Sigma, St Louis, MO, USA. All measurements of chemicals were standardized to 0.1 Molarity (M) ± 5% using an Ohaus Analytical Plus electronic balance accurate to within ± 0.0001 gram (g). Three trials per step were performed and recorded with the data presented here represents the average of that total data. All data collected fell within a statistically acceptable ± 5% (p=0.05) internal margin of variance [7] with no outliers. As a baseline, 0.1 g of T as ribothymidine (Figure 9) was added to 30 milliliters (mL) of sterile deionized water, pH 7.0, heated to 37° C and maintained for 30 minutes (min). A sample was then procured, placed in a crystal quartz cuvette, and its UV/Vis spectrum obtained (Figure 10).
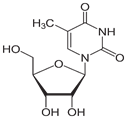
Figure 9: T as ribothymidine.
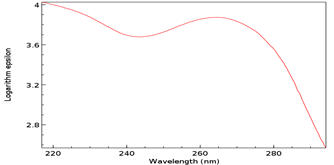
Figure 10: Baseline UV/Vis spectrum of T as ribothymidine. Note Absorbance peak of 3.9 at 267 nm.
To this mixture, 0.1 g of C as cytidine (Figure 11) and 0.1 g of SAM were added, with a resultant concentration of 0.3 g/30 mL or 1% and maintained at 37°C for 30 min. Concentration, pH, temperature, and time were all based on the aforementioned non-enzymatic DNA experiments [8]. A sample was then procured, placed in a crystal quartz cuvette, and its UV/Vis spectrum obtained (Figure 12).
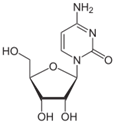
Figure 11: C as cytidine
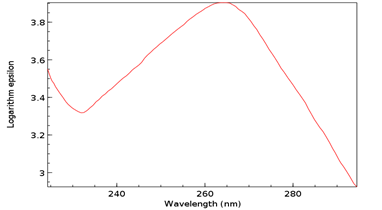
Figure 12: UV/Vis spectrum of T as ribothymidine following addition of C as cytidine and SAM. Note Absorbance peak of 4.1 at 267 nm.
3. Results and Discussion
The Absorbance peak of T as ribothymidine treated with C as cytidine and SAM at 4.1 versus that of the baseline at 3.9 represents a quantitative increase of 20% in the former according to the Beer-Lambert Law [9], far in excess of the 5% possible by chance. While any proposed base pairing between T as ribothymidine and the adenine moiety (A) of SAM would await future confirmation, Watson-Crick base-pairing rules would presuppose its likelihood and, although somewhat circuitous, upon base-pairing, the pathway from the non-enzymatic methylation of C as cytidine via SAM, first to the formation of 5-mC and then to T as ribothymidine via spontaneous deamination, may represent a rudimentary metabolism as the first instance of a prebiotically plausible molecule, T [10], utilizing other prebiotically plausible molecules, SAM [11] and C [12], to replicate itself, with SAH possibly being re-methylated to regenerate SAM via non-enzymatic methylation by membrane phosphatidylcholine (PC), similar to the non-enzymatic methylation of folic acid by PC [13], with PC itself potentially being prebiotic, as PC-like molecules have been detected in at least one meteorite [14]. Indeed, a primitive form of gene expression and/or silencing might be at work here as well, with both a phenotype (pyrimidine) and genotype (T) which may or may not be expressed, depending on whether 5-mC base pairs with G, halting deamination and thus T formation. Furthermore, the ideal RNA template sequence to facilitate such replication with fidelity would be the mRNA initiation sequence AUG, with UG ensuring it minimally (Figure 13). Such triplet nucleotide sequences have also been shown experimentally to act as templates for RNA-catalyzed RNA ribozymes when nucleotide triphosphates were used as substrates [15].
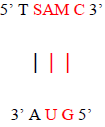
Figure 13: Ideal RNA template sequence to facilitate T self-replication with fidelity, with minimal sequence highlighted in red.
Additionally, base-pairing to the AUG template sequence may account for the conserved TΨC sequence in the T-loop section of the TΨC arm of tRNA, with Ψ representing pseudouridine (Figure 14), a modified base found in RNA, which base pairs with any other base [16], possibly accounting for the origin of this conserved sequence (Figure 15).
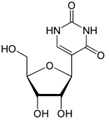
Figure 14: Pseudouridine
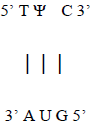
Figure 15: AUG RNA template for the conserved TΨC sequence of the T-loop section of the TΨC arm of tRNA.
The TΨC arm and T-loop are hypothesized to be the most ancient part of tRNA, which is itself the most modified and methylated nucleic acid, postulated to possibly be the most ancient surviving artifact of the RNA world, evolved from either two homologous halves or, more likely, three homologous minihelices, and while the T-loop in all but one archaea, the genus Pyrococcus, with archaea believed to be the most ancient of organisms [17], possesses the modified base N1 methyl pseudouridine (Figure 16) instead of T [18], pseudouridine is itself a prebiotically implausible molecule and may presuppose a role in some capacity for the more prebiotically plausible T [19], although a scenario could be envisaged in which N1 methyl pseudouridine feasibly supplants the role of T in the above described settings. Considering these factors, despite the SAM-dependent presence of T in its tRNA being attributed to ancient horizontal gene transfer from bacteria [20], the archaeal genus Pyrococcus could still be considered the likeliest candidate for the Last Universal Common Ancestor (LUCA) (Figure 17).
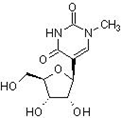
Figure 16: N1 methyl pseudouridine
Figure 17: Stacked Venn diagram representing the presence of T in the tRNA of Prokaryotes.
The experimentally verified non-enzymatic methylation of C by SAM to form 5-mC and its subsequent spontaneous deamination to T may very well represent, therefore, the first, or very close to the first, possibly by way of N1methyl pseudouridine, self-replicating molecule. Additionally, it may be the process responsible for selection of UG and AUG as the sequence for the mRNA translation initiation codons, the conserved TΨC sequence in the T-loop of the TΨC arm of tRNA, and methionine as the universally carried starting amino acid in proteins. Finally, the presence of T in an RNA world in which U was otherwise employed may have been part of the bridge to the present DNA-protein world in which T is favored over U.
4. Prospectus
Although the non-enzymatic methylation of C by SAM to 5-mC and its subsequent spontaneous deamination to T in DNA at physiologic pH and temperature has been implicated in some C to T point mutagenic cancers, cancers in general display a global hypomethylation of their DNA [21]; SAM, therefore, sold as the over-the-counter (OTC) supplement SAM-e, may still possibly play a role as a potential rescue adjunct therapy in certain cancers, particularly those treated with alkylating chemotherapeutic agents, much in the same way that folic acid is used as a rescue adjunct therapy when using antifolate chemotherapeutic agents [22]. Clinical trials in support of this prospectus are, therefore, justifiable.
Acknowledgements
I thank my fellow faculty and staff at Blue Ridge Community and Technical College for the generous use of their facilities and equipment. I also thank Dr. Zachary Burton, Professor Emeritus at Michigan State University (MSU) for invaluable insight and discussion, without whom this paper would not have been possible, and I especially thank my loving family for their generous gift of time in the performance of these experiments and the writing of this paper.
References
- Mazin AL, Gimadutdinov OA, Turkin SI, et al. Non-enzymatic DNA methylation by S-adenosylmethionine results in the formation of minor thymine residues and 5-methylcytosine from cytosine. Mol Biol (Mosk) 19 (1985): 903-914.
- Hancock, RL. Theoretical mechanisms for synthesis of carcinogen-induced embryonic proteins: XII mutational and non-mutational mechanism as subsets of a more general mechanism. Part A-Ethionine. Med Hypotheses 15 (1984): 323-331.
- Robertson MP, Joyce GF. The origins of the RNA world. Cold Spring Harb Perspect Biol 4 (2012): a003608.
- Belinky F, Rogozin IB, Koonin EV. Selection on start codons in prokaryotes and potential compensatory nucleotide substitutions. Scientific Reports 7 Article number 12422 (2017).
- Koplik R. Ultraviolet and visible spectrometry. Advanced strategies in food analysis.
- Black J. Microbiology. Prentice Hall (1993): 334.
- Bolton S. Pharmaceutical statistics (3rd). New York: Marcel Dekker (1997).
- Rydberg B, Lindahl T. Nonenzymatic methylation of DNA by the intracellular methyl group donor S-adenosyl-L-methionine is a potentially mutagenic reaction. EMBO J 1 (1982): 211-216.
- Beer-Lambert Law.
- Choughuley AS, Subbaraman AS, Kazi ZA, et al. A possible prebiotic synthesis of thymine: uracil-formaldehyde-formic acid reaction. Biosystems 9 (1977): 73-80.
- IFI CLAIMS Patent Services. S-adenosyl methionine regulation of metabolic pathways and its use in diagnosis and therapy. EP1221615A2 (2006).
- Biscans A. Exploring the emergence of RNA nucleosides and nucleotides on the early earth. Life 8 (2018): 57.
- Kowiatek BK. An additional, complementary mechanism of action for folic acid in the treatment of megaloblastic anemia. Journal of Biotechnology and Biomedicine 1 (2018): 021-027.
- Wirick S, Cody G, Flynn GJ, et al. Detection of a water soluble component of the Tagish Lake meteorite. Lunar and Planetary Science XXXVI (2005).
- Attwater J, Raguram A, Morgunov AS, et al. Ribozyme-catalysed RNA synthesis using triplet building blocks. Elife 15 (2018): 7.
- Kierzek E, Malgowska M, Lisowiec J, et al. The contribution of pseudouridine to stabilities and structure of RNAs. Nucleic Acids Res 42 (2014): 3492-3501.
- Kim Y, Kowiatek B, Opron K, et al. Type-II tRNAs and evolution of translation systems and the genetic code. Int J Mol Sci 22 (2018): 19.
- Wurm JP, Griese M, Bahr U, et al. Identification of the enzyme responsible for N1-methylation of pseudouridine 54 in archaeal tRNAs. RNA 18 (2012): 412-420.
- Dworkin JP. Attempted prebiotic synthesis of pseudouridine. Orig Life Evol Biosph 27 (1997): 345-355.
- Walbott H, Leulliot N, Grosjean H, et al. The crystal structure of Pyrococcus abyssi tRNA (uracil-54, C5)-methyltransferase provides insights into its tRNA specificity. Nucleic Acids Res 36 (2008): 4929-4940.
- Ehrlich M. DNA hypomethylation in cancer cells. Epigenomics 1 (2009): 239-259.
- Hagner N, Joerger M. Cancer chemotherapy: targeting folic acid synthesis. Cancer Manag Res 2 (2010): 293-301.


 Impact Factor: * 5.3
Impact Factor: * 5.3 Acceptance Rate: 75.63%
Acceptance Rate: 75.63%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 