Finding Drugs for Poxviruses: Targeting the L1 Protein
Alessandro Careglio
Department of Drug Science and Technology of the University of Turin, Italy
*Corresponding author: Alessandro Careglio, Department of Drug Science and Technology of the University of Turin, Frazione Santa Maria 92/a La Morra 12064 (CN) Italy.
Received: 01 December 2024; Accepted: 09 December 2024; Published: 10 January 2025
Article Information
Citation: Alessandro Careglio. Finding Drugs for Poxviruses: Targeting the L1 Protein. Fortune Journal of Health Sciences. 8 (2025): 26-34.
View / Download Pdf Share at FacebookAbstract
This study computationally compares natural compounds and synthetic drugs, particularly antivirals, against known and novel protein targets of Poxviruses using Cresset Flare(computational software). While several antiviral drugs have exhibited in vitro anti-poxvirus activity, cidofovir, a nucleoside analog approved for cytomegalovirus retinitis treatment in AIDS, has shown limited therapeutic utility due to its significant nephrotoxicity. The following computational approaches were employed:
• Ligand-based screening: Using cidofovir as a reference, a library of approximately 3000 natural compounds and common drugs, as well as a library of 700 antiviral molecules, were screened.
• Structure-based screening: A co-crystallized cidofovir-protein complex (PDB ID 5KM8) was used as a template to screen a library of natural compounds.
• Protein-protein docking: S. pneumoniae topoisomerases, for which drug-bound structures are available, were aligned with topoisomerases lacking ligands to identify potential binding sites.
• Covalent docking: Serine and tyrosine residues within hydrophobic pockets were targeted with drugs and natural compounds containing nitrile groups.
The poxvirus L1 protein, a conserved target across the poxvirus family, presents a myristoylated envelope protein with a hydrophobic cavity. This cavity is essential for virion assembly and represents a potential target for antibody-based therapies and vaccine development [5]. By identifying potential inhibitors for these critical poxvirus targets, this study aims to contribute to the development of novel antiviral strategies.
Keywords
<p>poxvirus, smallpox, computational chemistry, natural compounds</p>
Article Details
Introduction
Poxviruses, including the historically devastating smallpox virus, are among the largest viruses capable of infecting both vertebrates and invertebrates. Following the eradication of smallpox and the subsequent discontinuation of the vaccine in the early 1980s, interest in these viruses waned. Poxviruses are large, brick-shaped, double-stranded DNA viruses with complex symmetry. Their genomes encode proteins essential for viral replication, including DNA and RNA polymerases, kinases, and phosphatases. The core genes, conserved across poxviruses, are responsible for essential viral functions. In contrast, the terminal genes, which vary between different poxviruses, influence host range and virulence. This enables poxviruses to replicate within host cells, hijacking cellular processes for their own benefit [10].
Chordopoxviruses are epitheliotropic, typically causing skin lesions. Infections can range from mild, localized skin lesions to severe, systemic diseases like smallpox. While many poxvirus infections resolve spontaneously, severe cases or infections in immunocompromised individuals may require antiviral therapy [1]. Cidofovir, a nucleoside analog approved for cytomegalovirus retinitis, has shown some efficacy against poxviruses. However, its limited therapeutic use due to nephrotoxicity highlights the need for novel antiviral strategies. The L1 protein, a conserved component of the poxvirus envelope, represents a promising target for antiviral interventions. Its hydrophobic cavity plays a crucial role in virion assembly and could be targeted by antibody-based therapies or vaccine development [1].
Materials and Methods
Computational comparisons were performed using Flare from Cresset: ligand-based and structurebased screening, protein alignment, and covalent docking and screening on BLAZE servers.
The analyzed compounds were derived from two sources:
- • Selleckchem2 database: A source of synthetic antiviral compounds.
- • PubChem database: A source of natural compounds, including those from ivy, St. John's wort, cocoa, and natural nitrile compounds, downloaded in SDF format.
Three target proteins were used for the analysis:
- • Poxvirus L1 protein: A myristoylated envelope protein with a hydrophobic pocket essential for viral replication.
- • Smallpoisomerase: A DNA topoisomerase enzyme.
- • DNA topoisomerase (PDB ID 2L8P): A DNA topoisomerase structure from the Protein Data Bank.
Ligand-Based Alignment
Cidofovir Library vs. Natural Compounds
In this step, we identify molecules with the greatest similarity to cidofovir based on the electrostatic fields they generate. The comparison is performed against a database of natural compounds and drugs (selleckchem.com) containing approximately 2,600 entries. We utilize the SDF format for the compound library downloaded from the website.
Molecules with a similarity coefficient greater than 0.6 (considering an identity of 1) were selected.
Result: Several molecules exhibited a high degree of similarity to the reference compound, 2-Deoxycytidine 5-monophosphate, a drug used to treat leukemia. Among these, Cytidylic acid, 5-Methyl-2'-deoxycytidine, and sulforaphane (found in many Brassicaceae species) were identified. Extensive research has been conducted on the clinical use of sulforaphane, including its antiviral activity against various coronavirus variants. Additionally, 5Methylcytidine and cytidine were found to be similar to the reference compound.
Complete screening data: See Screenshot 1 for details. Refer to the caption for a description of the parameters.
Ligand-Based Alignment
A ligand-based screening was performed on a library of 700 antiviral compounds, using cidofovir as a reference.
Result: Lamivudine and encitabine were identified as the compounds with the greatest similarity to the reference molecule.
Complete screening data: See Screenshot 2 for details. Refer to the caption for a description of the parameters.
Structure-based Screening
DNA Docking (PDB ID 2L8P) with Antiviral Compounds A co-crystal structure of cidofovir bound to a DNA molecule (PDB ID 2L8P) is available. Cidofovir forms covalent bonds with the DNA molecule through its phosphate group and the hydroxyl group of DNA. By excluding cidofovir and performing non-covalent docking with the same ligand, similar binding poses were observed, although with lower scores (see Additional Figure 2). A detailed interpretation of the results can be found in Screenshot 3 and its caption.
Results: Several compounds exhibited promising docking scores:
- • Neferin: A nootropic compound with antiviral effects against SARS-CoV-2.
- • Nilotinib: An antineoplastic drug.
- • KIN 1148: A modulator of the antiviral immune response.
- • Camptothecin: An antineoplastic topoisomerase inhibitor.
- • Dynasore: An inhibitor of clathrin- and dynamin-dependent endocytosis.
- • Cellcept and Gandotinib: Drugs with potential antiviral activity.
Docking on DNA (PDB ID 2L8P) with a Natural Compound Library
The protein structure (PDB ID 2L8P) was prepared by removing the original ligand. This protein structure was then used to dock a library of natural compounds. (See Screenshot 4)
Results: The following compounds exhibited the best docking scores:
- • Neferin: A nootropic compound with antiviral properties.
- • Halofunginone: A coccidiostat used in veterinary medicine.
- • Magnoflorine: A sedative and anti-inflammatory compound.
The antiviral activity of neferin has been confirmed by Yang Yang et al. (9).
DNA Docking (PDB ID 2L8P) on BLAZE Server
The BLAZE server (Cresset) was used to screen compound libraries based on electrostatic and shape similarity to assess the likelihood of binding to the target protein. The cidofovirprotein complex (PDB ID 2L8P) was used as a reference. Approximately 150,000 compounds were screened against this reference.
Results: CHEMBL1213016 was identified as the compound with the greatest similarity to the reference molecule. However, this compound lacks a toxicological profile, biological activity data, and clinical use. Similarly, the first seven compounds ranked by the server did not meet these criteria.
Protein Alignment
The topoisomerases from S. pneumoniae (PDB ID 3RAE) and smallpox virus (PDB ID 3IGC) were aligned. This alignment was necessary because the crystallized smallpox protein lacks a reference drug or molecule to highlight potential binding pockets, unlike the S. pneumoniae topoisomerase, which is complexed with levofloxacin. The alignment revealed a sequence identity of 15.56% between the two proteins (Additional Figure 3). Unfortunately, significant structural similarities were not identified between the two proteins (Additional Figure 5).
Docking on Smallpox Topoisomerase (PDB ID 3IGC)
A docking study was performed on the smallpox topoisomerase using a library of compounds derived from cocoa (Theobroma cacao L., 1753) (Table 1). A grid encompassing the entire protein was used for the docking calculations.
The docked compounds exhibited affinity for two specific regions of the protein. Interestingly, these regions overlap with the binding sites of levofloxacin on S. pneumoniae topoisomerase, as shown in Additional Figure 6.(4)
Results: These compounds exhibited a high affinity for both topoisomerases. The structures of the catechin derivatives (catechin, epicatechin, gallocatechin (GC), epigallocatechin (EGC), catechin gallate, epicatechin gallate, gallocatechin gallate, and epigallocatechin gallate (EGCg)) are shown in Screenshot 5. These compounds demonstrated docking scores comparable to the co-crystallized ligand. As confirmed by Yoscida et al. (6), these catechin derivatives possess potential biological activity.(11)
Docking of St. John's Wort Compounds on Smallpox Topoisomerase (PDB ID 3IGC)
Smallpox infects subcutaneous areas, and Hypericum oil, with its active principles, could potentially reach these infected sites. Previous research by Katherine et al. (6) has confirmed the activity of Hypericum compounds against topoisomerases. Docking studies were performed on the smallpox topoisomerase using a library of compounds derived from St. John's Wort (Hypericum perforatum). Compounds such as procyanidin B2, pseudohypericine, mentoflavone, hypericine, isoquercitrin, and biapigenin exhibited high affinity for the topoisomerase, as indicated by the docking scores (see Screenshot 6).(12)
Covalent Docking on the Poxvirus L1 Protein
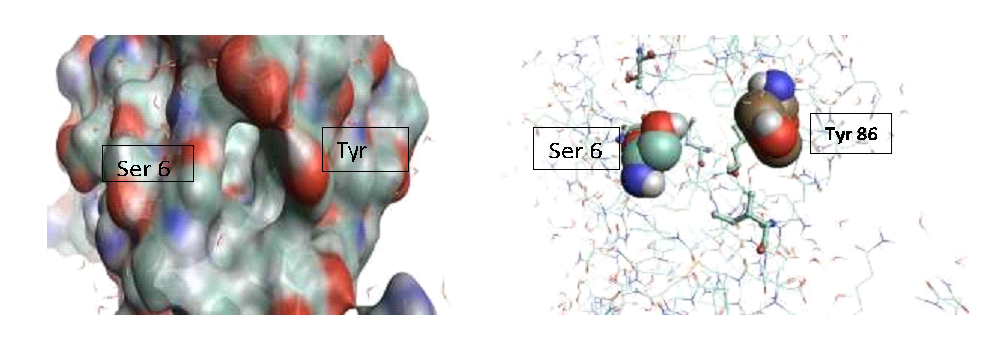
A library of natural compounds containing nitrile groups (Table 2) was docked onto the L1 protein of the poxvirus. The L1 protein is a conserved myristoylated envelope protein that plays a crucial role in virion assembly. It contains a hydrophobic cavity lined with 16 hydrophobic amino acids: Ile-7, Thr-10, Val-11, Leu-14, Ala-72, Thr-75, Tyr-76, Leu-79, Val-87, Met-90, Phe-91, Val-104, Phe-108, Leu-163, Leu-166, and Ala-170. This cavity is essential for shielding the myristoylated region of the protein.
Figure 11: represents the molecular surface of the L1 protein on the left and a stick representation on the right. Serine 6 and tyrosine 86 are highlighted at the entrance to the hydrophobic pocket. The myristoylated residue, essential for viral membrane assembly, would enter and exit through this pocket.(4)
Tyrosine 86 was chosen as a target residue for covalent binding with compounds containing nitrile groups.
The ligands to be attached must possess an appropriate functional group, also known as a covalent warhead (e.g., Michael acceptors, nitriles, alkynylamides, alpha-halo ketones). These electrophilic groups initially bind non-covalently to the target protein and then react with a specific nucleophilic residue (cysteine, lysine, serine, or tyrosine) at the active site, forming a covalent bond.
Results: Menisdaurin exhibited the highest score, considering electrostatic complementarity (EC Rho), as shown in Additional Figure 7. Vilazodone, a drug used to treat major depressive disorder, exhibited the highest score. It binds covalently to serine at the entrance of the pocket, occupying the site (Additional Figure 8). Other compounds with high scores include neratinib (a tyrosine kinase inhibitor and antitumor drug), BMS-191095 (a potassium channel opener), and epanolol (a betablocker).
Covalent Docking with Natural Compounds from Hops and Ivy on the Poxvirus L1 Protein
Several compounds from hops (Table 4) and ivy (Table 5) were docked onto the L1 protein to assess their potential for covalent binding to nucleophilic residues, such as tyrosine.
Results: Compounds like falcarinol, falcarinol, xantohumol, rosmarinic acid, vitamin C, and vitamin K3 exhibited high affinity for the L1 protein and were predicted to bind covalently to tyrosine residues. Adhumulon, while not forming a covalent bond, showed high electrostatic complementarity and could potentially bind non-covalently, followed by a slower covalent binding process. Covalent binding to the L1 protein could lead to its permanent inactivation. Please refer to Screenshot 8 for further details.
Conclusions
Despite the eradication of smallpox in the 1980s, recent cases have highlighted the need for effective antiviral treatments, especially considering the limited therapeutic options beyond cidofovir, which suffers from significant side effects. This study aimed to identify potential natural compounds and drugs that could combat smallpox infection. Several promising candidates were identified:
- • Approved Drugs:
o 2-Deoxycytidine 5-monophosphate (used for leukemia)
o Lamivudine and Encitabine (antiviral drugs)
o Neferin (nootropic with antiviral properties)
o Nilotinib (antineoplastic drug)
o KIN 1148 (immunomodulator)
o Camptothecin (topoisomerase inhibitor)
o Dynasore (endocytosis inhibitor)
o Vilazodone (antidepressant)
o Neratinib (tyrosine kinase inhibitor)
o BMS-191095 (potassium channel opener)
o Eponalol (beta-blocker) - • Natural Compounds:
o Sulforaphane (found in cruciferous vegetables)
o Alliin (found in garlic)
o Artemisic acid (found in Artemisia plants)
o Halofunginone (veterinary drug)
o Catechin derivatives (e.g., catechin, epicatechin, gallocatechin, epigallocatechin, and their gallate esters)
o Menisdaurin (a natural nitrile compound)
o Falcarinol, falcarinol, xantohumol, rosmarinic acid, vitamin C, vitamin K3, and adhumulone (found in various plants, including hops and ivy)
These compounds exhibited high affinity for viral proteins, particularly the L1 protein, and have the potential to inhibit viral replication through various mechanisms, including covalent binding to key residues. It would be desirable to use natural compounds in combination with drugs to reduce the adverse effects of drug therapies. Considering that plant metabolites can influence the activity of drugmetabolizing enzymes, they can either decrease or increase plasma drug concentrations, potentially leading to overdoses. In particular, Hypericum, based on the obtained scores, could be effective when used topically with Hypericum oil for treating lesions of the stratum corneum. It is important to note that Hypericum should not affect the activity of hepatic cytochromes. The grammar in that section is already correct, but here's a slightly improved version for better readability:
Conflicts of Interest
No conflicts of interest were declared.
Funding
This study was self-funded.
Acknowledgements
The author would like to thank Cresset for providing an academic license for Flare software.
Video Presentation
Statement on the use of Generative Artificial Intelligence
This manuscript has been translated and grammatically corrected using Gemini.
References
- N Carlone, R Pompei V. Microbiologia Farmaceutica Tullio ISBN 9788836230211
- http://www.selleckchem.com
- https://pubchem.ncbi.nlm.nih.gov
- Kimberly M Maize, Rachit Shah, Alex Strom, Sidath Kumarapperuma, Andrew Zhou, et al. A Crystal Structure Based Guide to the Design of Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) Activated Provides Finzel Mol Pharm 14 (2017): 3987-3997.
- Su HP, Garman SC, Allison TJ, Fogg C, Moss B, Garboczi DN. The 1,51-Å structure of the poxvirus L1 protein, a target of potent neutralizing antibodies. Proc Natl Acad Sci USA 102 (2005): 4240-5.
- Yoshida N, Kuriyama I, Yoshida H, Mizushina Y. Inhibitory effects of catechin derivatives on mammalian DNA polymerase and topoisomerase activities and mouse one-cell zygote development. J Biosci Bioeng 115 (2013): 303-9.
- A Peebles , Ronda K Baker , Ebba U Kurz , BJ Schneider , David J Kroll. Catalytic inhibition of human DNA topoisomerase IIα by hypericin, a naphthodianthrone from St. John’s wort (Hypericum perforatum).
- Cabras – Martelli (Piccin). Chimica degli Alimenti
- Protein Data Bank i) 3RAE Quinolone(Levofloxacin)-DNA cleavage complex of type IV topoisomerase from S. pneumoniae ii) 3IGC Smallpox virus topoisomerase-DNA transition state.
- Yang Yang, Peng Yang, Cong Huang, Yuming Wu, Zhe Zhou. Inhibitory effect on SARS-CoV-2 infection of neferin by blocking ca2+ -dependent membrane fusion. J Med Virol 93 (2021): 5825-5832.
- Kuzuhara T, Sei Y, Yamaguchi K, Suganuma M, Fujiki H. DNA and RNA as new binding targets of green tea catechins. J Biol Chem 23 (2006): 17446-17456.
- Veronika Butterweck, Mathias Schmidt.St. John's wort: role of active compounds for its mechanism of action and efficacy. Wien Med Wochenschr 157 (2007): 356-61.













 Impact Factor: * 6.2
Impact Factor: * 6.2 Acceptance Rate: 76.33%
Acceptance Rate: 76.33%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 