Study on the Relationship Between Adverse Events and Efficacy of Immunotherapy for Gastric Cancer and Target Prediction
Zidong Zhao1,2,#, Xu Zhang2,#, Yu Tan3, Dandan Zhao4, Satoshi Endo*,1,5,6, Yanwen Liu*,2,
1United Graduate School of Drug Discovery and Medical Information Sciences, Gifu University, Gifu 501-1194, Japan
2School of Medicine, Southeast University, Nanjing 210009, Jiangsu, China.
3Huangdao Traditional Chinese Medicine Hospital, Qingdao266500, Shandong Province, China 3. Department of Gastroenterology, the Affiliated Hospital of Xuzhou Medical University, Xuzhou 221002, Jiangsu Province, China.
4Huangdao Traditional Chinese Medicine Hospital, Qingdao 266500, Shandong Province, China
5Center for One Medicine Innovative Translational Research (COMIT), Institute for Advanced Study, Gifu University, Gifu 501-1193, Japan
6Preemptive Food Research Center, Institute for Advanced Study, Gifu University, Gifu 501-1193, Japan
#These authors contributed equally to this work.
*Corresponding Author Satoshi Endo, United Graduate School of Drug Discovery and Medical Information Sciences, Gifu University, Gifu 501-1194, Japan.
Yanwen Liu, School of Medicine, Southeast University, Nanjing 210009, Jiangsu, China.
Received: 14 October 2025; Accepted: 18 October 2025; Published: 21 October 2025
Article Information
Citation: United Graduate School of Drug Discovery and Medical Information Sciences, Gifu University, Gifu 501-1194, Japan
View / Download Pdf Share at FacebookAbstract
Objective: This study aimed to dissect molecular mechanisms of immune-related adverse events (irAEs) using tumor genomics data. We screened key regulatory pathways via immune enrichment analysis and identified core gene biomarkers through integrated bioinformatics and machine learning approaches, providing a basis for early irAE warning, efficacy evaluation, and personalized treatment.
Methods: RNA sequencing data of gastric adenocarcinoma (TCGA-STAD) were retrieved from TCGA. Six irAE-related immune pathways (Neutrophils, T cell receptor [TCR], Eosinophils, etc.) were selected. Pathway activity was validated by GSVA and GSEA. Key genes were screened and their diagnostic value evaluated using UpSet analysis, Boruta algorithm, and XGBoost model.
Results: The six pathways were significantly enriched in high irAE score samples, with Neutrophils (NES=1.97), TCR (NES=1.96), and Eosinophils (NES=1.84) showing highest enrichment. Five key genes (C11orf21, RGS19, etc.) were identified. RGS19 was highly expressed in gastric cancer tissues with optimal diagnostic performance (AUC=0.799).
Conclusion: Six irAE-related pathways (Neutrophils, TCR, etc.) and five key genes (including C11orf21, RGS19) were identified from TCGA. RGS19, highly expressed in gastric cancer with prominent diagnostic efficacy, provides clinical and molecular evidence for irAE prediction and early diagnosis.
Keywords
<p>Gastric Cancer; Immune-Related Adverse Events (irAEs); Target prediction; Enrichment analysis</p>
Article Details
Introduction
Currently, the clinical incidence of gastric cancer remains persistently high in China, with most patients diagnosed at an advanced stage. As a major disease threatening human life and health, the initiation and progression of malignant tumors are closely associated with the abnormal proliferation of tumor tissues and the immunosuppressive state of the body [1]. Tumor cells can evade immune system surveillance through multiple mechanisms, among which the activation of immune checkpoint pathways is one of the key approaches[2]. Driven by in-depth research in immuno-oncology, immune checkpoint inhibitors (ICIs) have emerged. These agents block immunosuppressive signaling pathways to reactivate anti-tumor immune responses, thereby achieving immune-mediated clearance of tumor cells [3]. Although the clinical application of ICIs has significantly enhanced anti-tumor immune effects, the accompanying immune-related adverse events (irAEs) still require high clinical attention. Gaining in-depth insights into the mechanisms underlying irAEs, and enabling their early identification and standardized intervention, are crucial components for optimizing the management of tumor immunotherapy. While immunomodulatory therapy activates anti-tumor immune responses, it may also induce irAEs due to the overactivation of autoimmune reactions. ICIs may impair the tolerance of peripheral blood to self-antigens and induce the production of autoantibodies—a mechanism closely associated with the development of irAEs in different organs [4-6]. IrAEs share significant similarities with autoimmune diseases; numerous clinical case reports have shown that ICIs can induce substantial immune responses resembling those of autoimmune diseases, suggesting that irAEs may represent the overt manifestation of subclinical autoimmune reactions in some patients.
Although the clinical characteristics of irAEs have been widely recognized, their exact pathophysiological mechanisms remain incompletely elucidated. The academic community has proposed multiple explanatory mechanisms, including autoantibody production, T cell infiltration, and mediation by inflammatory cytokines such as interleukins (ILs) [7]. Current research suggests that the occurrence of irAEs is directly related to changes in the function of the body’s autoimmune system, primarily manifested as the disruption of autoimmune tolerance mechanisms, or the enhanced sensitivity of the body to antigen recognition (which leads to attacks on its own tissues) [5]. IrAEs can affect various organ systems throughout the body; common affected systems include the skin, digestive system, endocrine system, and respiratory system, while rare cases may present with neurotoxicity and cardiotoxicity [8].Currently, the treatment of irAEs mainly relies on corticosteroids, immunosuppressants, and cytokine blockers. However, these therapies may suppress the immune system function of patients, thereby weakening anti-tumor immune responses. At present, irAE research still faces many unresolved issues, such as unclear mechanisms of occurrence, lack of specific biomarkers, difficulties in early identification, and absence of precise personalized treatment regimens [9]. Previous clinical applications of ICIs in tumor types such as lung cancer and melanoma have accumulated extensive experience in irAE management, but relevant research data in gastric cancer remain relatively scarce. Meanwhile, clinically effective predictive indicators for irAEs are lacking. Known potential risk factors (e.g., tumor gene mutations, autoantibodies, and gut microbiota) mostly rely on specialized laboratory tests. Due to high requirements for testing conditions and low reproducibility, the clinical promotion and application of these tests are significantly limited. Therefore, exploring predictive indicators of irAEs with clinical application value is of great significance for guiding clinical treatment decisions and formulating personalized treatment regimens.
Methods
Enrichment Analysis of irAE-Related Immune Pathways
RNA-seq data (initial FPKM format) and clinical profiles of gastric adenocarcinoma (TCGA-STAD) were retrieved from TCGA. FPKM was converted to TPM for cross-sample comparability, then normalized via log2(TPM + 1); samples with complete data formed the analysis cohort. Six irAE-related immune pathways (Eosinophils, Neutrophils, TNF, IFN, TCR, TMB) were identified: Eosinophils/Neutrophils gene sets from GeneCards (by functional relevance), others from prior studies. GSVA quantified pathway activity (unsupervised, no pre-set thresholds), and GSEA verified pathway enrichment by analyzing gene set distribution in ranked genome-wide expression lists.
Intersection Screening of irAE Pathway-Associated Genes
Spearman correlation analysis (suitable for non-normal biological data) quantified associations between each gene’s expression and pathway GSVA scores. UpSet intersection analysis visualized overlapping genes with positive correlations (cor > 0) across all 6 pathways, identifying key genes with multi-pathway regulatory roles.
Feature Importance Evaluation of Key Genes
Boruta (random forest-based) assessed the diagnostic value of intersection genes: "shadow features" (randomly permuted gene data) were trained alongside original genes. Genes outperforming their shadow features in importance scores were deemed significant for irAE classification.
Multi-Model Comparison for Optimal Diagnostic Model
TCGA-STAD samples were split 6:4 into training/test sets (balanced irAE-positive/negative ratios). Nine machine learning algorithms (Logistic regression, LDA, FDA, MDA, RF, SVM, NB, GBM, XGBoost) built irAE diagnostic models. Performance was evaluated via 10-fold cross-validation; average AUC (primary metric, 0.5 = random, 1.0 = perfect classification) determined the optimal model (XGBoost).
SHAP-Based Model Interpretation & Visualization
SHAP (from game theory) interpreted the XGBoost model: SHAP values quantified each gene’s marginal contribution to predictions (positive = promotes irAE prediction, negative = inhibits). SHAP dependence plots (non-linear gene-prediction relationships) and summary plots (global gene importance) visualized results.
Key Gene Expression & Diagnostic Value
Five key genes (C11orf21, FUT7, KCNJ10, RGS19, ZNF101) were analyzed: Wilcoxon rank-sum tests compared their expression in gastric adenocarcinoma vs. normal tissues (TCGA-STAD data). ROC curves and AUC values assessed diagnostic efficacy (AUC closer to 1.0 = better discrimination).
Results
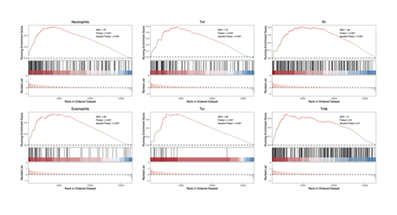
Pathways may play critical roles in the development of immune-related adverse events (irAEs). To explore the potential role of immune-related features in the occurrence of irAEs, this study analyzed samples from the TCGA-GC cohort. First, gene set variation analysis (GSVA) was performed to obtain pathway enrichment scores for each sample. Samples were then grouped based on these scores, followed by gene set enrichment analysis (GSEA). In the analysis of immune-related pathways, six pathways—Neutrophils, Tnf, Ifn, Eosinophils, TCR, and Tmb—were focused on. The results showed that all six pathways were significantly enriched in samples with high irAE scores. Specifically, the normalized enrichment score (NES) is a key indicator for measuring the degree of pathway enrichment: the NES of the Neutrophils pathway reached 1.97, the TCR pathway was 1.96, and the Eosinophils pathway was 1.84; the NES values of the remaining Tnf, Ifn, and Tmb pathways were all greater than 1.3. Meanwhile, in terms of statistical significance, all P-values were less than 0.01, and the adjusted P-values also confirmed the significant statistical significance of the enrichment (Figure 1). The above results strongly indicate that the immune features corresponding to these immune-related pathways are highly likely to play key roles in the development of irAEs. Among them, the Neutrophils, TCR, and Eosinophils pathways, due to their more prominent enrichment levels, may exert particularly important functions in the mechanism of irAE occurrence. This provides valuable research evidence at the immune-related pathway level for further deciphering the pathogenesis of irAEs and identifying potential intervention targets.
This figure shows the enrichment status of 6 pathways (Neutrophils, Tnf, Ifn, Eosinophils, TCR, and Tmb) in the TCGA-GC cohort samples grouped by high and low GSVA scores. It includes the normalized enrichment score (NES), P-value, and adjusted P-value, all of which indicate that the enrichment has significant statistical significance. These results reflect that the relevant immune features may play a key role in the occurrence of immune-related adverse events (irAEs).
C11orf21, FUT7, KCNJ10, and Other Genes Are Intersection Genes of irAE-Related Pathways
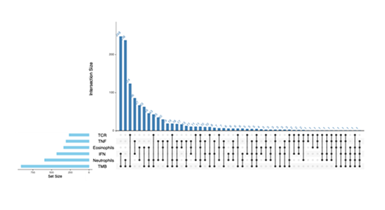
In the study of the regulatory mechanisms between immune-related pathways and immune-related adverse events (irAEs), we systematically investigated the correlation between genes and pathway scores. The analysis revealed that each of the six immune-related pathways—TCR (T-cell receptor pathway), TNF (tumor necrosis factor pathway), Eosinophils (eosinophil-related pathway), IFN (interferon pathway), Neutrophils (neutrophil-related pathway), and TMB (tumor mutational burden-related pathway)—contains a large number of genes that are positively correlated with the scores (correlation > 0). This finding lays a foundation for identifying key genes involved in cross-pathway regulation. To accurately identify core genes that are commonly involved in regulation across multiple pathways, we performed an UpSet intersection analysis on the gene sets with positive correlation (cor > 0) from the six aforementioned pathways. From the visualization results, the bar section of the UpSet plot shows the size of intersections among different gene sets, while the dot-line structure below indicates the participation of each pathway’s gene set in the intersections. The analysis results demonstrated that there are five genes that exhibit positive correlation across all six pathways, namely C11orf21, FUT7, KCNJ10, RGS19, and ZNF101 (Figure 2).
The upper bar chart shows the size of intersections among different gene sets (Intersection Size), reflecting the number of genes that are simultaneously involved in the positively correlated gene sets of multiple pathways. The lower dot-and-line plot presents the participation of gene sets from each pathway (TCR, TNF, Eosinophils, IFN, Neutrophils, TMB) in the intersections. This figure facilitates the identification of genes that play a common role across multiple pathways and helps explore key node genes regulating the occurrence of immune-related adverse events (irAEs).
Validation of Diagnostic Efficacy of Key Genes via Boruta Algorithm
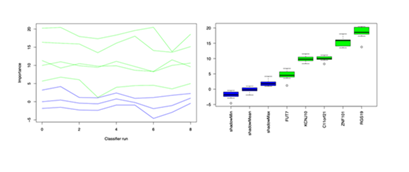
To clarify the diagnostic efficacy of the 5 candidate genes (C11orf21, FUT7, KCNJ10, RGS19, ZNF101) screened by UpSet intersection analysis for sample identification, this study conducted in-depth evaluation using the Boruta algorithm. From the algorithm’s result visualization (Figure 3): In the left panel (feature importance score trajectory plot), the 5 candidate genes (green lines) showed slight fluctuations in importance scores during 0–8 classifier runs, but their overall scores remained significantly higher than those of shadow variables (blue lines). In some runs, the peak importance score of candidate genes approached 20, while shadow variables mostly stayed in the low range or even showed negative values—highlighting the candidate genes’ superiority in feature contribution. In the right panel (variable importance boxplot), all 5 genes (green boxes) were classified as "Confirmed" features. Their importance distribution (taking RGS19 as an example, where the box and whiskers reflect central tendency and dispersion) was much higher than that of shadow variables (blue boxes). In conclusion, validation via the Boruta algorithm demonstrated that C11orf21, FUT7, KCNJ10, RGS19, and ZNF101 exhibit stable and reliable diagnostic efficacy in sample identification. This finding lays a solid molecular foundation for subsequent exploration of their roles in the irAE regulatory network and the development of precise diagnosis and intervention strategies.
Left panel: Trajectory of importance scores for real genes (green) and shadow variables (blue) across different classifier runs; Right panel: Variable importance boxplot (green boxes = Confirmed important features, blue boxes = shadow variables), intuitively demonstrating the stable and reliable diagnostic ability of the candidate genes.
Accurate Prediction of irAE-Related Features by XGBoost Model
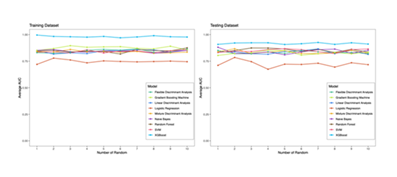
Nine machine learning models—Flexible Discriminant Analysis, Gradient Boosting Machine, Linear Discriminant Analysis, Logistic Regression, Mixture Discriminant Analysis, Random Forest, SVM, and XGBoost—were validated via 10 random samplings in the training and testing datasets (x-axis: random sampling ID; y-axis: average AUC).
As shown in Figure 4, XGBoost exhibited significant advantages in multiple validation rounds: its average AUC was consistently the highest among all models with small fluctuations, indicating good stability. In contrast, the other 8 models (e.g., Linear Discriminant Analysis, Logistic Regression) had lower overall average AUC and more obvious curve fluctuations (even if their AUC was close to XGBoost in some rounds), showing poor stability in classification performance. This confirms that the XGBoost model can more accurately capture data patterns and output reliable classification results in learning and predicting irAE-related features, laying an algorithmic foundation for subsequent diagnostic model construction.
The left and right panels respectively show the changes in average AUC values of 9 models across 10 random samplings in the training and testing datasets, intuitively demonstrating the advantages of the XGBoost model in classification performance and stability.
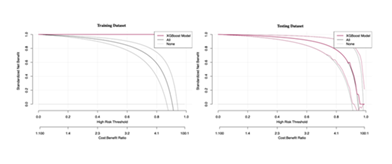
Clinical Benefit Evaluation of XGBoost Model via Decision Curve Analysis (DCA)
To clarify the practical clinical utility of the XGBoost model, decision curve analysis (DCA) was performed to assess its standardized net benefit across different high-risk thresholds. As shown in Figure 5, in both training and testing datasets, the XGBoost model’s DCA curve significantly outperformed the "treat-all" (labeled "All" in the figure) and "treat-none" (labeled "None" in the figure) strategies: within most high-risk threshold ranges, XGBoost yielded higher net benefits. For instance, when the cost-benefit ratio was within the clinically common range of 1:100 to 100:1, XGBoost’s standardized net benefit remained consistently higher than the other two strategies as the high-risk threshold changed. This indicates that in practical clinical decision-making, diagnostic results based on the XGBoost model can help clinicians more reasonably identify high-risk patients requiring intervention, avoid "over-treatment" or "missed diagnosis/treatment," and provide strong support for clinical decisions.
The left and right panels correspond to the training and testing datasets, respectively, showing the model’s standardized net benefit across different risk thresholds and demonstrating its clinical decision-making value compared with the "treat-all" and "treat-none" strategies.
Combining the AUC results from multi-model comparison and clinical value validation via DCA, the XGBoost model demonstrated the optimal classification performance and stability in both the training and testing datasets. Furthermore, decision curve analysis confirmed its promising potential for guiding clinical decision-making. Therefore, this study ultimately determined to adopt the XGBoost algorithm for constructing the irAE-related diagnostic model. This model is expected to provide an efficient and reliable tool for early identification, accurate diagnosis of irAEs, and the formulation of clinical intervention strategies, thereby promoting the optimization and improvement of the irAE diagnosis and treatment process.
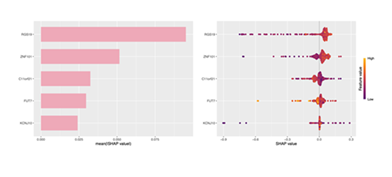
Confirmation of Strong Association Between Gene Expression and irAE Risk Prediction via SHAP Analysis
After constructing the irAE diagnostic model and identifying XGBoost as the optimal algorithm, SHAP (SHapley Additive exPlanations) was used to analyze the contribution and impact patterns of the 5 candidate genes (C11orf21, FUT7, KCNJ10, RGS19, ZNF101) in the model, enhancing biological interpretability for subsequent functional validation and clinical translation. From the mean SHAP value bar chart (Figure 6, left), the global contribution of genes in XGBoost differed significantly: RGS19 had the highest mean |SHAP value| (most prominent contribution), followed by ZNF101 (stronger influence than C11orf21/FUT7), while KCNJ10 showed the lowest contribution. This gradient clarified the hierarchical roles of genes in the model. The SHAP scatter plot (Figure 6, right; color = gene expression: orange = high, purple = low; x-axis = SHAP value: positive = promotion, negative = inhibition) further revealed expression-risk associations: RGS19/ZNF101 (major risk drivers): High expression (orange points) correlated with significantly increased positive SHAP values, strongly promoting irAE risk prediction.C11orf21/FUT7 (moderate risk-associated genes): High expression showed mild positive SHAP values, assisting in risk assessment as auxiliary factors.KCNJ10 (potential protective factor): Most points fell in the negative SHAP region; low expression (purple points) enhanced negative influence, potentially reducing irAE risk. SHAP analysis confirmed the gene contribution ranking (RGS19 > ZNF101 > C11orf21 ≈ FUT7 > KCNJ10) and their expression-based risk regulatory patterns. This converts the XGBoost “black box” into interpretable genetic logic, guiding subsequent functional validation (e.g., exploring RGS19/ZNF101’s pro-disease mechanisms) and clinical translation (using these genes as diagnostic biomarkers/intervention targets for irAE precision medicine).
Bar chart of mean SHAP values for each gene, quantifying their global contribution to the model. Right panel: SHAP scatter plot (point color = gene expression level; x-axis = impact direction/intensity on prediction), illustrating gene expression-irAE risk associations.
Accurate Prediction of irAE-Related Features by the XGBoost Model
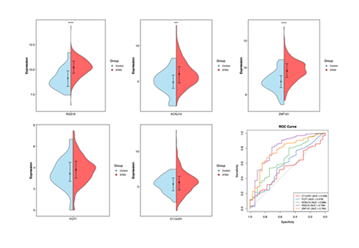
After clarifying the mechanism of the 5 candidate genes (C11orf21, FUT7, KCNJ10, RGS19, ZNF101) in the XGBoost diagnostic model, gene expression difference analysis and ROC curve validation were conducted to verify their expression characteristics and diagnostic potential in gastric adenocarcinoma (STAD) clinical samples.
Violin plot analysis (Figure 7) showed that except for C11orf21, RGS19, KCNJ10, ZNF101, and FUT7 were significantly upregulated in STAD samples (red distribution) compared with normal tissues (blue distribution) (all P<0.001): RGS19, KCNJ10, ZNF101, and FUT7 exhibited concentrated high expression in STAD (red regions shifted to high-expression intervals with distinct mean lines), while C11orf21 showed overlapping expression distributions and no significant difference between the two groups. In conclusion, RGS19, KCNJ10, ZNF101, and FUT7 showed significant expression features in STAD (upregulated except C11orf21), with RGS19 (AUC=0.799) demonstrating outstanding diagnostic value. This validates the effectiveness of machine learning-screened genes and supports their clinical translation: RGS19 could be developed into a core diagnostic marker, while ZNF101/KCNJ10 may form a multi-gene panel to improve accuracy. Additionally, these results guide further exploration of gastric cancer pathogenesis and precision medicine development.
(Show expression distribution differences of RGS19, KCNJ10, ZNF101, FUT7, and C11orf21 (normal tissues = blue; STAD samples = red), illustrating gene upregulation/downregulation trends and statistical significance. Bottom-right panel (ROC curves): Quantify diagnostic performance of each gene in distinguishing STAD from normal tissues via AUC values, supporting their clinical application potential.)
Discussion
Programmed death-1 (PD-1)/Programmed death-ligand 1 (PD-L1) monoclonal antibodies, as novel clinical antibody-based anti-tumor immunotherapeutics, have been widely used in the treatment of various malignancies, including colorectal cancer, non-small cell lung cancer, gastric cancer, and esophageal cancer. In the current field of tumor therapy, the combination of immunotherapy with conventional strategies (such as targeted therapy, chemotherapy, chemoradiotherapy) and dual immunotherapy has become a common clinical tumor treatment model. In-depth research on the clinical efficacy of immunotherapy in patients with gastroesophageal cancer and the immune-related adverse events (irAEs) occurring during treatment not only provides an important basis for exploring new clinical pathways of immunotherapy combined with targeted therapy for malignant tumors but also offers new insights for optimizing anti-tumor treatment strategies. Against the backdrop of continuous iteration and advancement of anti-tumor treatment regimens, an increasing number of optimized combined anti-tumor treatment regimens are gradually being applied in clinical practice, bringing more treatment options and survival hope to tumor patients. In this study, analysis of the TCGA-STAD cohort revealed that six immune-related pathways were significantly enriched in samples with high irAE scores, among which the Neutrophils, T cell receptor (TCR), and Eosinophils pathways showed the highest enrichment levels, with normalized enrichment scores (NES) of 1.97, 1.96, and 1.84, respectively.
This result is consistent with the previous theory that abnormal activation of immune cells is involved in the occurrence of irAEs. Excessive activation of neutrophils may exacerbate tissue damage by releasing pro-inflammatory factors[10], while abnormal activation of the TCR pathway may enhance the ability of T cells to recognize self-antigens, triggering autoimmune responses. The role of eosinophils in allergic and autoimmune diseases has been widely reported[11], and their high enrichment in irAEs suggests that they may be involved in tissue damage by releasing cytotoxic granular proteins. These findings provide a new perspective for in-depth understanding of the immunopathological mechanism of irAEs. In addition, through intersection analysis of the gene sets of the six pathways, this study identified five key genes that were positively correlated in all pathways: C11orf21, FUT7, KCNJ10, RGS19, and ZNF101. Boruta algorithm analysis showed that all five genes had stable diagnostic discriminative ability, suggesting that they may play key regulatory roles in the occurrence and development of irAEs. Among them, RGS19 had the highest contribution in the XGBoost model, and its high expression was significantly associated with increased irAE risk. As a regulator of G protein signaling, RGS19 may be involved in the activation and migration of immune cells by regulating G protein-coupled receptor signaling pathways[12]. ZNF101, a member of the zinc finger protein family, may be involved in the regulation of immune responses by regulating gene transcription[13]. FUT7 is involved in fucosylation modification and may affect the expression of cell surface antigens and immune recognition[14].
KCNJ10, as a potassium channel protein, may affect the function of immune cells by regulating intracellular electrolyte balance[15]. The specific functions of these genes and their mechanisms of action in irAEs require further experimental verification. Finally, this study compared the predictive performance of nine machine learning models and found that the XGBoost model exhibited the optimal classification performance in both the training and testing sets (with the highest average AUC). Decision curve analysis showed that its net benefit was superior to the "treat-all" and "treat-none" strategies in most risk threshold ranges. This result indicates that the XGBoost model constructed based on the five key genes has good clinical decision support value and is expected to be used for irAE risk prediction. SHAP analysis further revealed the contribution of each gene: RGS19 and ZNF101 were major risk drivers, while KCNJ10 may have a protective effect. These findings not only enhance the biological interpretability of the model but also provide a theoretical basis for the clinical development of irAE prediction biomarkers.
This study still has some limitations. First, the study data were obtained from the TCGA database, and all samples were from gastric adenocarcinoma patients; thus, the applicability of the results in other cancer types needs to be verified. Second, this study only conducted analysis based on gene expression data, lacking verification at the protein level and functional experiments, and the specific mechanism of action of key genes remains unclear. In addition, the study did not consider the impact of clinical factors (such as patients' immunotherapy regimens and baseline immune status) on the occurrence of irAEs. In conclusion, through enrichment analysis of irAE-related immune pathways, this study identified five key genes and constructed an efficient XGBoost prediction model, providing an important basis for understanding the molecular mechanism of irAEs and developing clinical prediction tools. Future studies need to further verify and improve these findings to promote the development of precise management of irAEs.
Conclusion
This study focused on immune-related adverse events (irAEs) associated with gastric cancer immunotherapy. Based on gastric adenocarcinoma data from The Cancer Genome Atlas (TCGA), six irAE-related immune pathways were screened, including neutrophils and T cell receptor (TCR) pathways. Among these, the Neutrophils, TCR, and Eosinophils pathways showed the highest enrichment levels, with normalized enrichment scores (NES) of 1.97, 1.96, and 1.84, respectively. Five key genes were identified through gene intersection analysis. Following validation via the Boruta algorithm and model construction using XGBoost, genes such as RGS19 were confirmed as major risk drivers for irAEs. These genes exhibited high expression in gastric cancer tissues and demonstrated excellent diagnostic efficacy—specifically, RGS19 achieved an area under the curve (AUC) of 0.799. Collectively, this study provides molecular biomarkers for irAE prediction, laying a foundation for the clinical application of irAE risk assessment in gastric cancer immunotherapy.
Declarations of Conflict of interest
No conflicts of interest are declared by the authors.
Funding
This study was supported by the funding from the National Health Commission Science and Technology Development Research Center, Major Program for Four Major Chronic Diseases (2023ZD0501500).
Consent for publication
All authors consented for the publication. No identifying images or other personal or clinical details of participants are presented that compromise anonymity.
Author Contributions
Zidong Zhao (first author) and Xu Zhang (co-first author) were responsible for data analysis and manuscript writing. Yu Tan and Dandan Zhao participated in data analysis and manuscript revision. Satoshi Endo and Yanwen Liu (corresponding authors) designed the research project, revised the manuscript, and gave final approval. All authors have read and approved the final manuscript.
References
- Eggermont AMM, et al. Association Between Immune-Related Adverse Events and Recurrence-Free Survival Among Patients With Stage III Melanoma Randomized to Receive Pembrolizumab or Placebo: A Secondary Analysis of a Randomized Clinical Trial. JAMA Oncol 6 (2020): 519-527.
- Verschoor YL, et al. Neoadjuvant atezolizumab plus chemotherapy in gastric and gastroesophageal junction adenocarcinoma: the phase 2 PANDA trial. Nat Med 30 (2024): 519-530.
- Andre T, et al. Neoadjuvant Nivolumab Plus Ipilimumab and Adjuvant Nivolumab in Localized Deficient Mismatch Repair/Microsatellite Instability-High Gastric or Esophagogastric Junction Adenocarcinoma: The GERCOR NEONIPIGA Phase II Study. J Clin Oncol 41 (2023): 255-265.
- Hasan Ali O, et al. Characterization of nivolumab-associated skin reactions in patients with metastatic non-small cell lung cancer. Oncoimmunology 5 (2016): e1231292.
- Oliveira C, et al. Immune-related serious adverse events with immune checkpoint inhibitors: Systematic review and network meta-analysis. Eur J Clin Pharmacol 80 (2024): 677-684.
- Wan G, et al. Multi-organ immune-related adverse events from immune checkpoint inhibitors and their downstream implications: a retrospective multicohort study. Lancet Oncol 25 (2024): 1053-1069.
- Sullivan RJ, JS Weber. Immune-related toxicities of checkpoint inhibitors: mechanisms and mitigation strategies. Nat Rev Drug Discov 21 (2022): 495-508.
- Dougan M, et al. Understanding and treating the inflammatory adverse events of cancer immunotherapy. Cell 184 (2021): 1575-1588.
- Chen CY, et al. Comparative safety of immune checkpoint inhibitors and chemotherapy in advanced non-small cell lung cancer: A systematic review and network meta-analysis. Int Immunopharmacol 108 (2022): 108-848.
- Li Huihui. Self-recruited neutrophils trigger excessive activation of innate immune responses and phenotypic transformation of cardiomyocytes in fulminant myocarditis (2024).
- Li Xiaofang and Zhang Yingmin. Mendelian randomization study on the correlation between bronchial asthma, peripheral eosinophil count, and allergic or autoimmune diseases. Journal of Environment and Health 41 (2024): 1076-1081.
- Hwang J, et al. RGS19 converts iron deprivation stress into a growth-inhibitory signal. Biochem Biophys Res Commun 464 (2015): 168-175.
- Ansari A, et al. Casz1 and Znf101/Zfp961 differentially regulate apolipoproteins A1 and B, alter plasma lipoproteins, and reduce atherosclerosis. JCI Insight 10 (2025).
- Cui Hongxia, et al. Effect of overexpression of exogenous FUT7 on the adhesion and migration abilities of breast cancer cells. Cancer Prevention and Treatment Research 41 (2014): 252-255.
- Zhang Xiaodong, Shi Jing, Li Weirong. Association between KCNJ10 gene rs6690889 single-nucleotide polymorphism and susceptibility to drug-resistant epilepsy in adults. Chinese Journal of Drugs and Clinical Medicine 21 (2021): 256-257.


 Impact Factor: * 6.2
Impact Factor: * 6.2 Acceptance Rate: 76.33%
Acceptance Rate: 76.33%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 