Seroprevalence & Molecular Characterisation of Hepatitis-C Virus Among High-Risk Group Cases Attending to a Tertiary Care Hospital of Eastern India
Manaswini Das1, Dharitri Mohapatra2, Sonam Silpa Moharana3, Roma Rattan4, Nirupama Chayani5, Sanatan Behera*,6
1Senior Resident, Microbiology, SCBMCH, Cuttack, Odisha, India
2Professor & HOD, Microbiology, SCBMCH, Cuttack, Odisha, India
3Assistant professor, Microbiology, MJKMCH, Jajpur, Odisha, India
4Associate Professor, Biochemistry, SCBMCH, Cuttack, Odisha, India
5Exprofessor & HOD, Microbiology, SCBMCH, Cuttack, Odisha, India
6Associate Professor, Hepatology, SCBMCH, Cuttack, Odisha, India
*Corresponding author: Sanatan Behera, Associate Professor, Hepatology, SCBMCH, Cuttack, Odisha, India.
Received: 05 August 2025; Accepted: 20 August 2025; Published: 10 September 2025
Article Information
Citation: Manaswini Das, Dharitri Mohapatra, Sonam Silpa Moharana, Roma Rattan, Nirupama Chayani, Sanatan Behera. Seroprevalence & Molecular Characterisation of Hepatitis-C Virus Among High-Risk Group Cases Attending to a Tertiary Care Hospital of Eastern India. Fortune Journal of Health Sciences. 8 (2025): 852-860.
View / Download Pdf Share at FacebookAbstract
Aim: To determine the seroprevalence and viral load in high-risk group patients and assess their compliance after treatment for Hepatitis C virus (HCV) infection.
Method: This hospital-based prospective study was conducted at SCB Medical College and Hospital, Cuttack from December 2019 to November 2021. A total of 200 high-risk group patients' blood samples were tested for HCV using Standard Q HCV antibody test (ICT), commercially available 3rd generation Microlisa (ELISA), and HCV RNA detection via Truenat real-time RT-PCR machine based on the 5'NCR region. Viral load monitoring after treatment was performed, and genotyping was conducted using the cobas 4800 system.
Results: Out of 200 high-risk group cases, 25% were ICT positive, 22.5% were ELISA positive, and 15% were HCV RNA positive. Among all high-risk groups, hemodialysis cases had the highest prevalence rate, and healthcare workers were significantly (p<0.05) associated with HCV infection. Genotype 3 was more predominant (68.5%) than genotype 1. Among genotype 1, subtype 1A (17.5%) was more prevalent than 1B (8.5%).
Conclusion: To rule out false positive cases, HCV RNA testing should be performed for all positive cases via Truenat real-time RT-PCR machine, which is portable, less time-consuming, requires minimal technical expertise, and is available at peripheral centres. This approach is also helpful for viral load monitoring before and after treatment.
Keywords
<p>HCV, viral load, hemodialysis, ELISA, genotyping</p>
Article Details
Introduction
Hepatitis C virus (HCV) is a small enveloped, single positive-stranded RNA virus from the genus Hepacivirus, family Flaviviridae, with a diameter of 50-60 nm [1,2]. Currently, HCV infection is primarily acquired through long-term hemodialysis, percutaneous blood exposure, intravenous drug abuse, and blood transfusion procedures such as those required for thalassemia patients [3,4]. It represents the common cause of post-transfusion hepatitis in developing countries and poses a significant global health challenge. According to the World Health Organization, Hepatitis C virus infects 2-3% of the world's population, corresponding to approximately 130-170 million individuals, with more than 350,000 people dying each year from HCV-related complications [6,7]. The prevalence in India is estimated to be about 0.5-1.5%, affecting approximately 12-18 million people [8]. This substantial burden necessitates comprehensive screening and management strategies, particularly for high-risk populations. Molecular techniques play a crucial role in HCV diagnosis and management as they not only help detect HCV RNA but also confirm the active state of infection, indicating that the virus is in a replicating state within the patient's body [9,10]. In immunocompromised individuals, enzyme-linked immunosorbent assay (ELISA) may yield false-negative results, making molecular testing the preferred choice for accurate detection [14]. The identification of populations with high-risk behaviour through anti-HCV ELISA followed by confirmatory RT-PCR molecular testing for viral load estimation before and after treatment provides valuable insights into therapeutic response and helps in early identification and treatment decisions.
The heterogeneity of HCV presents unique challenges in terms of treatment and prevention strategies. After determination of the infecting HCV genotype, patients with chronic HCV infection have specific treatment options available [11,12]. It is generally accepted that HCV genotype determination and genotype 1 subtype classification (1a/1b) are essential assessments prior to treatment initiation, as different genotypes respond differently to various therapeutic regimens [13]. The clinical presentation of HCV infection varies widely, ranging from asymptomatic cases to severe manifestations including chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma [15,16]. High-risk groups such as hemodialysis patients, individuals requiring frequent blood transfusions, intravenous drug users, and healthcare workers are particularly vulnerable to HCV infection due to their increased exposure to potentially contaminated blood and blood products [17,18]. Keeping these considerations in view, the present study was conducted at SCB Medical College and Hospital with high-risk group patients including those undergoing hemodialysis, requiring blood transfusions, and intravenous drug abusers, both symptomatic and asymptomatic individuals. The study aimed to comprehensively evaluate the seroprevalence of HCV infection in these populations and characterize the molecular aspects of the virus to inform better diagnostic and treatment strategies.
Material and Methods
Study Design and Place of Study
This study was designed as a hospital-based prospective study conducted in the viral research diagnostic laboratory, Department of Microbiology, and molecular genomic laboratory, Department of Biochemistry, SCB Medical College and Hospital, Cuttack, Odisha. The study utilized a comprehensive approach combining serological and molecular techniques to achieve accurate diagnosis and characterization of HCV infection.
Study Period
The present study was carried out over a period of 2 years from December 2019 to November 2021. This extended duration allowed for adequate sample collection and comprehensive analysis of seasonal variations and treatment outcomes in the study population.
Study Population
The study was conducted on all high-risk group cases, including both outpatients and inpatients admitted to SCB Medical College and Hospital, Cuttack. The high-risk groups included persons co-infected with HIV, injecting drug users, hemodialysis patients, thalassemia patients requiring repeated blood transfusions, and individuals presenting with fever, jaundice, and abdominal pain. Patients with hepatitis B virus (HBV), hepatitis A virus (HAV), hepatitis E virus (HEV) infections, and non-infectious hepatitis cases were excluded from the study to maintain specificity and avoid confounding factors that could affect the interpretation of results.
Sample Collection
From the high-risk group cases, 5 ml of venous blood was collected into BD vacutainer tubes following standard phlebotomy procedures. Proper patient identification and sample labelling were ensured throughout the collection process to maintain sample integrity and traceability.
Sample Processing
The blood collected was separated into two vials for different analytical purposes. Three millilitres of blood were taken for rapid diagnostic testing and ELISA in clot activator vials, while another 2 ml of blood was collected in EDTA vials for HCV RNA processing. All samples were appropriately labelled and numbered, with details entered into standardized data collection forms. In the clot activator vial, blood was allowed to stand for 15 minutes to ensure complete clot formation. Subsequently, the samples were centrifuged at 6000 rpm for 15 minutes, and the clear serum supernatant was carefully transferred to plain vials. One millilitre of serum was allocated for immunochromatographic testing (RDT), while another 2 ml was reserved for ELISA processing. Samples were stored at -80°C until further processing to maintain sample stability and prevent degradation.
Standard Q HCV Ab Test
The Standard Q HCV Ab test is a rapid test that utilizes chromatographic and immunoassay systems to detect HCV antibodies through a rapid immunoassay technique that provides the qualitative detection of antibodies against HCV. It represents a rapid system to enable the detection of specific IgG antibodies in human serum, plasma and whole blood samples for HCV.
The test is based upon principles established by the SD BIOSENSOR STANDARD Q HCV Ab test containing pre-coated two lines nitrocellulose membrane in the control and test region. These are non-visible as the sample adds. Two monoclonals react to anti-NS3 and anti-core, anti-IgG human coated anti-hummon monoclonals are used to coat the control line whereas on test/strip as well in region T and C of the nitrocellulose membrane. Two hundred nanomeric HCV recombinant antigens, from core and NS3 domain, conjugation to colloidal gold particles is used for the detection of the antibody complex. In this procedure, sample-specific antigens present on complex HCV interact with Colloidal gold-HCV antigens which forms antigen-antibody gold particle complex This complex, through capillary action moving through the membrane for the test the complex will get caught via monoclonal anti-human IgG antibody leading to a visible violet colour on the TEST line on HCV Antibody present.
Enzyme-Linked Immunosorbent Assay (ELISA)
A human serum and/or whole blood or plasma 4th gen ELISA immunoassay-based sandwich for detecting antibodies against HCV (anti-HCV) were investigated with in-vitro qualitative. The 3rd-generation Microlisa assay for Ig G against HCV uses antigens including antigen, which has specific epitopes for capturing both IgM-class anti-HCVs and IgG-class anti-cores, C-20 and D-43 specific epitopes for anti-core C. HCV-3-generation Microlisa shows better specificity compared to second or first-generation ELISA systems. The microwells are initially coated with antigens, structural and non-structural parts from CORE to NS4 and antigens derived from NS5 region, and diluted analytes together with controls for assaying.
Truenat HCV
Truenat HCV is an in situ hybridization test, (chip) real-time reverse transcription (RT) PCR technology for qualitative detection of quantitative Hepatitis C virus RNA in human plasma or serum and whole blood: An easy-to-use, real-time PCR solution for decentralized and near-patient HCV monitoring with accurate viral load values in single-wavelength technology, simple operation, no need of technical training and good robustness The technology is based on the real-time Reverse Transcriptase Polymerase-Che Chain Reaction (RT-PCR) Taqman based system. The HCV genome 5prime NS2 / NS3 /4region and the 3´UTR region is also targeted by primers and are detected for specific primer region (Target Amplicon size: ≈ 73bp) of target sequence is not required in PCR, primer oligo-nuelectrode probes used in Cobas system.
Genotyping
It is a qualitative nucleic acid amplification (NAAT) based Cobas HCV v 2.0 assay performed on the Cobs 4800 System. Real- time reverse transcription-polymerase chain reaction (RT-PCR) on target amplification is used on the cobas 4800 System to identify Genotypes 1 to 6 and Sub-type 1a and 1b through two-step amplification using genotype- and subtype-specific primers fluorescent dye labelled oligo probes. Also detects the HCV does not genotype related using primers and probes targeting a high degree conserved element in the viral genome to use as a control sample in total pre-PCR and RT-PCR processes (internal control). This is a qualitative test that detects genomes when it is present in all genotypes using genus-specific primers and a fluorescent labelled oligonucleotide probe with an internal control for an entire test.
Ethical Consideration
The study protocol titled “Seroprevalence and Molecular Characterisation of Hepatitis C Virus Among High-Risk Group Cases Attending a Tertiary Care Hospital of Eastern India” underwent review and was approved by the Institutional Ethics Committee (IEC) of S.C.B. Medical College and Hospital, Cuttack (IEC Regd. No. ECR/84/Inst/OR/2013/RR-20; IEC Application No. 144). Expedited reviews took place on 29 January 2020 and 5 February 2020, followed by a full committee review at the 36th IEC meeting held on 7 February 2020. Approval was granted for a period of three years, with the requirement to report any adverse events and obtain IEC approval for any protocol amendment
Results
The present study included 200 high-risk group cases with equal distribution of hemodialysis and blood transfusion cases (25% each), as shown in Figure 1. The remaining cases comprised chronic liver disease patients (15%), intravenous drug abusers (15%), and healthcare workers (20%).
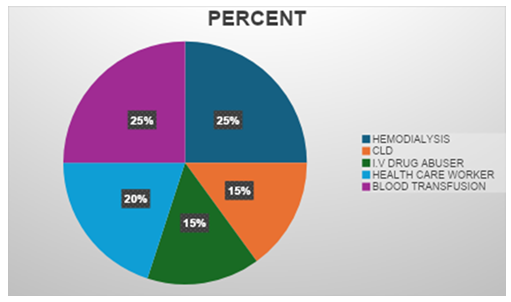
Figure 1: Distribution of high-risk group cases included in the study (n=200). The study population comprised equal proportions of hemodialysis patients and blood transfusion recipients (25% each), followed by healthcare workers (20%), chronic liver disease patients (15%), and intravenous drug abusers (15%).
As presented in Table 1, out of 200 high-risk group cases, rapid diagnostic testing (RDT) showed 50 positive cases (25%), ELISA demonstrated 45 positive cases (22.5%), and RT-PCR confirmed 30 positive cases (15%). This pattern indicates RDT positivity > ELISA positivity > HCV RNA positivity, suggesting the presence of false-positive results in serological testing methods.
|
Method |
Positive Cases (n) |
Positivity Rate (%) |
|
RDT |
50 |
25 |
|
ELISA |
45 |
22.5 |
|
RT-PCR (HCV RNA) |
30 |
15 |
|
Total Screened |
200 |
— |
Table 1: Comparison of HCV detection rates using different diagnostic methods among 200 high-risk group cases. RT-PCR showed the lowest positivity rate (15%) compared to serological methods (RDT: 25%, ELISA: 22.5%), indicating the presence of false-positive results in antibody-based testing.
Table 2 demonstrates RDT positivity among different high-risk groups. Hemodialysis patients showed the highest positivity rate at 40% (20/50), followed by chronic liver disease patients at 26.66% (8/30), blood transfusion recipients at 24% (12/50), intravenous drug abusers at 20% (6/30), and healthcare workers at 10% (4/40).
|
HIGH RISK GROUP |
RDT POSITIVITY |
PERCENTAGE |
|
Hemodialysis (50) |
20 |
40% |
|
Chronic liver disease (30) |
8 |
26.66% |
|
I.V drug abuser (30) |
6 |
20% |
|
Health care worker (40) |
4 |
10% |
|
Blood transfusion (50) |
12 |
24% |
|
Total (200) |
50 |
25% |
Table 2: Rapid diagnostic test (RDT) positivity rates across different high-risk groups. Hemodialysis patients demonstrated the highest positivity rate (40%), while healthcare workers showed the lowest rate (10%).
Seropositivity results, as shown in Table 3, revealed similar patterns with hemodialysis patients demonstrating the highest prevalence at 30% (15/50), followed by chronic liver disease patients at 26.66% (8/30), blood transfusion recipients at 24% (12/50), intravenous drug abusers at 20% (6/30), and healthcare workers at 10% (4/40).
|
HIGH RISK GROUP |
SEROPOSITIVITY/ELISA |
PERCENTAGE |
|
Hemodialysis (50) |
15 |
30% |
|
Chronic liver disease (30) |
8 |
26.66% |
|
I.V drug abuser (30) |
6 |
20% |
|
Health care worker (40) |
4 |
10% |
|
Blood transfusion (50) |
12 |
24% |
|
Total (200) |
45 |
22.50% |
Table 3: ELISA seropositivity rates among high-risk groups. Similar to RDT results, hemodialysis patients had the highest prevalence (30%), followed by chronic liver disease patients (26.66%).
HCV RNA positivity among high-risk groups, presented in Table 4, showed hemodialysis patients with the highest prevalence at 24% (12/50), followed by chronic liver disease patients at 20% (6/30), blood transfusion recipients at 14% (7/50), intravenous drug abusers at 10% (3/30), and healthcare workers at 5% (2/40).
|
HIGH RISK GROUP |
RNA POSITIVITY |
PERCENTAGE |
|
Hemodialysis (50) |
12 |
24% |
|
Chronic liver disease (30) |
6 |
20% |
|
I.V. drug abuser (30) |
3 |
10% |
|
Health care worker (40) |
2 |
5% |
|
Blood transfusion (50) |
7 |
14% |
|
Total (200) |
30 |
15% |
Table 4: HCV RNA positivity rates determined by RT-PCR across different high-risk groups. Hemodialysis patients maintained the highest prevalence (24%), confirming active viral replication in this population.
Statistical analysis using chi-square test, as shown in Table 5, revealed that healthcare workers had significantly lower prevalence (p<0.05) compared to other high-risk groups, with a prevalence of only 10% versus 25.6% in other groups combined.
|
HIGH RISK GROUP |
Elisa positive |
percentage |
X 2 |
PREVALENCE |
|
HEMODIALYSIS (50) |
15 |
30% |
2.151 |
0.14 |
|
CHRONIC LIVER DISEASE (30) |
8 |
26.66% |
0.351 |
0.55 |
|
IV DRUG ABUSER (30) |
6 |
20% |
0.127 |
0.72 |
|
HEALTH CARE WORKER (40) |
4 |
10% |
4.48 |
0.03 |
|
BLOOD TRANSFUSION (50) |
12 |
24% |
0.08 |
0.76 |
Table 5: Statistical analysis of HCV prevalence among high-risk groups using chi-square test. Healthcare workers showed significantly lower prevalence (p<0.05) compared to other high-risk groups.
The diagnostic performance evaluation showed that RDT had 100% sensitivity and 88.2% specificity when compared with HCV RNA results (Table 6). Similarly, ELISA demonstrated 100% sensitivity and 91.1% specificity (Table 7), indicating that both serological methods are highly sensitive but less specific than molecular testing.
|
RNA +VE |
RNA –VE |
|
|
RDT +VE |
30 |
20 |
|
RDT –VE |
0 |
150 |
Table 6: Diagnostic performance of rapid diagnostic test (RDT) compared to HCV RNA detection by RT-PCR. The RDT demonstrated 100% sensitivity and 88.2% specificity for detecting HCV infection.
Sensitivity = TP / (TP + FN) = 30 / (30 + 0) = 30 / 30 = 100%
Specificity = TN / (FP + TN) = 150 / (20 + 150) = 150 / 170 ≈ 88.2%
|
RNA +VE |
RNA −VE |
Total |
|
|
ELISA +VE |
30 |
15 |
45 |
|
ELISA −VE |
0 |
155 |
155 |
|
Total |
30 |
170 |
200 |
Table 7: Diagnostic performance of ELISA compared to HCV RNA detection by RT-PCR. ELISA showed 100% sensitivity and 91.2% specificity, indicating superior specificity compared to RDT.
Sensitivity = TP / (TP + FN) = 30 / (30 + 0) = 30 / 30 = 100%
Specificity = TN / (TN + FP) = 155 / (155 + 15) = 155 / 170 ≈ 91.2%
Laboratory findings among high-risk group patients (Table 8) revealed altered liver function tests as the most common abnormality (40%), followed by altered renal function tests and decreased haemoglobin levels (25% each). Other significant findings included hypoalbuminemia and prolonged prothrombin time/INR (20% each), and leucocytosis (15%).
|
LABORATORY PROFILE |
FREQUENCY |
PERCENT |
|
ALTERED LFT |
80 |
40% |
|
ALTERED RFT |
50 |
25% |
|
HYPOALBUMINEMIA |
40 |
20% |
|
LEUCOCYTOSIS |
30 |
15% |
|
PT/INR |
40 |
20% |
|
DECREASED HAEMOGLOBIN |
50 |
25% |
Table 8: Laboratory abnormalities observed in high-risk group patients (n=200). Altered liver function tests were the most common finding (40%), followed by altered renal function tests and decreased hemoglobin levels (25% each).
Gender distribution analysis showed male predominance with 76.5% (153/200) males and 23.5% (47/200) females among high-risk groups. Age-wise distribution demonstrated that the 51-60 years age group had the highest prevalence at 34.5% (69/200), followed by the 41-50 years age group at 19% (38/200). Clinical presentation analysis (Table 9) revealed that fatigue, nausea, and loss of appetite were the most common symptoms, affecting 85% (170/200) of patients. Other significant presentations included abdominal pain and ascites (25%), bleeding disorders such as melena and hematemesis (20%), fever and pruritus (15% each), and neurological symptoms including weight loss, confusion, drowsiness, and slurred speech (12.5%).
|
Clinical features |
Frequency |
percent |
|
Fatigue, Nausea, Loss of Appetite |
170 |
85% |
|
Fever |
30 |
15% |
|
Pruritus |
30 |
15% |
|
Abdominal pain & Ascites |
50 |
25% |
|
Bleeding disorders like Malena & Haematemesis |
40 |
20% |
|
Weight loss, Confusion, Drowsiness, Slurring of speech |
25 |
12.50% |
Table 9: Clinical presentations observed in high-risk group patients. Non-specific symptoms including fatigue, nausea, and loss of appetite were the most common presentations (85%), highlighting the insidious nature of HCV infection.
Associated complications showed jaundice as the most common complication affecting 15% (30/200) of patients, followed by liver cirrhosis in 5% (10/200), hepatocellular carcinoma in 2.5% (5/200), and hepatic encephalopathy in 1% (2/200) of cases. HIV co-infection analysis revealed that 1% (2/200) of high-risk group patients were HIV positive, with one case being HCV positive and another HCV negative. Therefore, HIV co-infection among HCV-positive patients was 3.3% (1/30). Treatment outcome evaluation showed that out of 30 HCV RNA-positive cases, 26 patients (86.7%) recovered successfully, while 4 cases (13.3%) did not recover due to poor treatment compliance. Genotyping results (Table 10) demonstrated that genotype 3 was the most prevalent, accounting for 68.5% (137/200) of cases, followed by genotype 1A at 17.5% (35/200), genotype 1B at 8.5% (17/200), genotype 4 at 3.5% (7/200), and indeterminate genotypes at 2% (4/200).
|
Genotype |
Prevalence |
Percentage (%) |
|
3 |
137 |
68.5 |
|
1A |
35 |
17.5 |
|
1B |
17 |
8.5 |
|
4 |
7 |
3.5 |
|
Indeterminate |
4 |
2 |
Table 10: HCV genotype distribution among the study population (n=200). Genotype 3 was the most prevalent (68.5%), followed by genotype 1A (17.5%) and genotype 1B (8.5%), reflecting the typical genotype distribution pattern in the Indian subcontinent.
Discussion
The present study provides comprehensive insights into the seroprevalence and molecular characteristics of hepatitis C virus infection among high-risk populations in Eastern India. The findings demonstrate significant variations in HCV prevalence across different risk categories and highlight the importance of molecular confirmation for accurate diagnosis. The overall seroprevalence of 22.5% by ELISA and confirmed viremia of 15% by RT-PCR in our study population indicates a substantial burden of HCV infection among high-risk groups. These findings are consistent with previous studies from similar healthcare settings in India, where high-risk populations demonstrate elevated HCV prevalence rates compared to the general population [19,20]. The significantly higher prevalence among hemodialysis patients (30% by ELISA, 24% by RT-PCR) aligns with established knowledge regarding nosocomial transmission risks in dialysis units. The chronic nature of hemodialysis treatment, frequent vascular access procedures, and potential for cross-contamination create an environment conducive to HCV transmission [21,22]. These findings emphasize the critical importance of strict infection control measures, regular screening protocols, and isolation procedures for HCV-positive patients in dialysis facilities. Chronic liver disease patients demonstrated the second-highest prevalence (26.66% by ELISA, 20% by RT-PCR), which is expected given that many patients with chronic liver disease may have underlying viral hepatitis as an etiological factor. The progression from acute to chronic infection and the potential for viral persistence in immunocompromised individuals contribute to this high prevalence [23].
Blood transfusion recipients showed considerable HCV positivity (24% by ELISA, 14% by RT-PCR), highlighting ongoing challenges in blood safety despite improvements in screening procedures. While the implementation of nucleic acid testing has significantly reduced transfusion-transmitted infections, the window period and potential for variant strains may still pose risks [24,25]. The relatively lower prevalence among intravenous drug users (20% by ELISA, 10% by RT-PCR) in our study may reflect the specific population characteristics and harm reduction programs in the region. However, this group remains at high risk due to needle sharing practices and associated high-risk behaviours [26]. Healthcare workers demonstrated the lowest prevalence (10% by ELISA, 5% by RT-PCR), which was statistically significant (p<0.05). This finding suggests that occupational safety measures and awareness programs may be effective in reducing transmission risks among healthcare personnel. However, continued vigilance and adherence to universal precautions remain essential [27]. The discrepancy between serological positivity and molecular confirmation (22.5% vs. 15%) underscores the importance of RT-PCR testing for definitive diagnosis. False-positive serological results can occur due to cross-reactivity, non-specific binding, or resolved infections where antibodies persist despite viral clearance [28]. Our study's demonstration of 100% sensitivity for both RDT and ELISA confirms their utility as screening tools, while their specificities of 88.2% and 91.1%, respectively, emphasize the need for molecular confirmation.
The predominance of genotype 3 (68.5%) in our study population is consistent with the genotype distribution pattern observed in the Indian subcontinent. Genotype 3 is known to be associated with more rapid progression to steatosis and fibrosis, making early detection and treatment particularly important [29,30]. The presence of genotype 1A (17.5%) and 1B (8.5%) reflects the genetic diversity of HCV in the region and has important implications for treatment selection, as different genotypes respond variably to direct-acting antiviral therapies. The clinical presentation findings reveal that most patients (85%) presented with non-specific symptoms such as fatigue, nausea, and loss of appetite, which emphasizes the insidious nature of HCV infection. The relatively low prevalence of jaundice (15%) and other specific hepatic manifestations suggests that many cases may remain undiagnosed without systematic screening of high-risk populations [31]. Laboratory abnormalities, particularly altered liver function tests (40%), provide important diagnostic clues and monitoring parameters. The presence of renal dysfunction (25%) among patients, particularly in the hemodialysis group, reflects the complex interplay between chronic kidney disease and viral hepatitis [32]. The treatment outcome analysis revealing an 86.7% success rate is encouraging and reflects the efficacy of modern direct-acting antiviral therapies. However, the 13.3% treatment failure rate due to poor compliance highlights the importance of patient education, support systems, and potentially simplified treatment regimens to improve adherence [33]. The male predominance (76.5%) observed in our study may reflect gender-related differences in risky behaviour patterns, occupational exposures, or healthcare-seeking behaviour. The higher prevalence in the 51-60 years age group suggests that many infections may have been acquired decades earlier when awareness and prevention measures were less developed [34]. The low HIV co-infection rate (3.3% among HCV-positive patients) is favourable compared to some high-risk populations in other regions, but continued surveillance and integrated care approaches remain important for managing co-infected patients [35].
Our study's utilization of the Truenat platform for HCV RNA detection demonstrates the feasibility of deploying molecular diagnostic technologies in resource-limited settings. The portability reduced technical complexity, and faster turnaround time of this system make it particularly suitable for decentralized testing approaches, potentially improving access to confirmatory diagnosis in peripheral healthcare facilities [36]. The comprehensive genotyping data generated through the cobas 4800 system provides valuable epidemiological information and supports personalized treatment approaches. Understanding local genotype distribution patterns is crucial for public health planning, treatment guideline development, and monitoring potential changes in viral population dynamics over time [37]. Several limitations of our study should be acknowledged. The hospital-based design may introduce selection bias, as patients seeking healthcare may have different characteristics compared to the general high-risk population. The cross-sectional nature of serological and molecular testing provides a snapshot of infection status but may not capture the dynamic nature of viral replication and immune responses. Additionally, the study period coinciding with the COVID-19 pandemic may have influenced healthcare-seeking behaviour and sample collection procedures. Future research directions should include longitudinal follow-up studies to assess treatment outcomes, viral resistance patterns, and long-term clinical progression. Community-based screening programs could provide more representative prevalence data and identify asymptomatic cases that might not seek healthcare. Integration of HCV screening with other routine healthcare services could improve case detection and linkage to care. The implementation of point-of-care testing strategies using rapid molecular platforms could revolutionize HCV diagnosis and management in resource-limited settings. Cost-effectiveness analyses of different screening and confirmation strategies would inform policy decisions and resource allocation priorities.
Conclusion
This comprehensive study demonstrates a significant burden of hepatitis C virus infection among high-risk populations attending a tertiary care hospital in Eastern India. The seroprevalence of 22.5% by ELISA and confirmed viremia of 15% by RT-PCR highlight the substantial disease burden in these vulnerable populations. Hemodialysis patients emerged as the highest-risk group with 30% seroprevalence, followed by chronic liver disease patients at 26.66%. The discrepancy between serological and molecular testing results emphasizes the critical importance of RT-PCR confirmation for accurate diagnosis, particularly in clinical decision-making and treatment initiation. To rule out false-positive cases, HCV RNA testing should be performed for all serologically positive cases using advanced molecular platforms such as the Truenat real-time RT-PCR system, which offers portability, reduced time requirements, minimal technical expertise needs, and availability at peripheral healthcare facilities. The predominance of genotype 3 (68.5%) followed by genotype 1A (17.5%) provides valuable epidemiological insights and supports the development of region-specific treatment guidelines. The successful treatment outcome in 86.7% of cases demonstrates the efficacy of current therapeutic approaches, while the 13.3% failure rate due to poor compliance underscores the need for enhanced patient support and adherence strategies.
Healthcare workers showed significantly lower HCV prevalence (p<0.05), suggesting that occupational safety measures and awareness programs may be effective in reducing transmission risks. However, continued vigilance and strict adherence to universal precautions remain essential across all healthcare settings. The study's findings support the implementation of systematic screening programs for high-risk populations, particularly hemodialysis patients, blood transfusion recipients, and individuals with chronic liver disease. The integration of rapid molecular diagnostic platforms can facilitate timely diagnosis, appropriate treatment initiation, and effective public health interventions. Future efforts should focus on developing comprehensive prevention strategies, improving diagnostic accessibility through point-of-care testing, enhancing treatment adherence support systems, and establishing robust surveillance mechanisms to monitor epidemiological trends and treatment outcomes. The successful implementation of these measures could significantly reduce the burden of hepatitis C infection and its associated complications in high-risk populations throughout Eastern India and similar healthcare settings globally.
Acknowledgements
The authors sincerely thank the Departments of Microbiology and Biochemistry at SCBMCH, Cuttack, and MJKMCH, Jajpur, for their support to conduct this study.
Conflicts Of Interest
The authors affirm that there are no commercial, financial, or personal relationships that could be construed as potential conflicts of interest in relation to this research.
References
- Choo QL, Kuo G, Weiner AJ, et al. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244 (1989): 359-362.
- Lindenbach BD, Rice CM. The ins and outs of hepatitis C virus entry and assembly. Nat Rev Microbiol 11 (2013): 688-700.
- Alter MJ. Epidemiology of hepatitis C virus infection. World J Gastroenterol 13 (2007): 2436-2441.
- Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis 5 (2005): 558-567.
- Lavanchy D. Evolving epidemiology of hepatitis C virus. Clin Microbiol Infect 17 (2011): 107-115.
- World Health Organization. Global health sector strategy on viral hepatitis 2016-2021. Geneva WHO (2016).
- Stanaway JD, Flaxman AD, Naghavi M, et al. The global burden of viral hepatitis from 1990 to 2013: findings from the Global Burden of Disease Study 2013. Lancet 388 (2016): 1081-1088.
- Sievert W, Altraif I, Razavi HA, et al. A systematic review of hepatitis C virus epidemiology in Asia, Australia and Egypt. Liver Int 31 (2011): 61-80.
- Pawlotsky JM. Use and interpretation of virological tests for hepatitis C. Hepatology 36 (2002): S65-73.
- Chevaliez S, Pawlotsky JM. Hepatitis C virus serologic and virologic tests and clinical diagnosis of HCV-related liver disease. Int J Med Sci 3 (2006): 35-40.
- Zeuzem S, Buti M, Ferenci P, et al. Efficacy of 24 weeks treatment with peginterferon alfa-2b plus ribavirin in patients with chronic hepatitis C infected with genotype 1 and low pretreatment viremia. J Hepatol 44 (2006): 97-103.
- Manns MP, McHutchison JG, Gordon SC, et al. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358 (2001): 958-965.
- Simmonds P, Bukh J, Combet C, et al. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42 (2005): 962-973.
- Lok AS, Gunaratnam NT. Diagnosis of hepatitis C. Hepatology 26 (1997): 48S-56S.
- Seeff LB. Natural history of chronic hepatitis C. Hepatology 36 (2002): S35-46.
- El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology 142 (2012): 1264-1273.
- Jadoul M, Poignet JL, Geddes C, et al. The changing epidemiology of hepatitis C virus (HCV) infection in haemodialysis: European multicentre study. Nephrol Dial Transplant 19 (2004): 904-909.
- Henderson DK. Managing occupational risks for hepatitis C transmission in the health care setting. Clin Microbiol Rev 16 (2003): 546-568.
- Mukhopadhya A. Hepatitis C in India. J Biosci 33 (2008): 465-473.
- Puri P, Anand AC, Saraswat VA, et al. Consensus statement of HCV task force of the Indian National Association for Study of the Liver (INASL). Part I: Status report of HCV infection in India. J Clin Exp Hepatol 4 (2014): 106-116.
- Fabrizi F, Takkouche B, Lunghi G, et al. The impact of hepatitis C virus infection on survival in dialysis patients: meta-analysis of observational studies. J Viral Hepat 14 (2007): 697-703.
- Jadoul M, Barril G. Hepatitis C in hemodialysis: epidemiology and prevention. Contrib Nephrol 176 (2012): 35-41.
- Hoofnagle JH. Course and outcome of hepatitis C. Hepatology 36 (2002): S21-29.
- Stramer SL, Glynn SA, Kleinman SH, et al. Detection of HIV-1 and HCV infections among antibody-negative blood donors by nucleic acid-amplification testing. N Engl J Med 351 (2004): 760-768.
- Biswas R, Tabor E, Hsia CC, et al. Comparative sensitivity of HBV NATs and HBsAg assays for detection of acute HBV infection. Transfusion 43 (2003): 788-798.
- Nelson PK, Mathers BM, Cowie B, et al. Global epidemiology of hepatitis B and hepatitis C in people who inject drugs: results of systematic reviews. Lancet 378 (2011): 571-583.
- Beltrami EM, Williams IT, Shapiro CN, Chamberland ME. Risk and management of blood-borne infections in health care workers. Clin Microbiol Rev 13 (2000): 385-407.
- Richter SS. Laboratory assays for diagnosis and management of hepatitis C virus infection. J Clin Microbiol 40 (2002): 4407-4412.
- Kumar D, Farrell GC, Fung C, George J. Hepatitis C virus genotype 3 is cytopathic to hepatocytes: Reversal of hepatic steatosis after sustained therapeutic response. Hepatology 36 (2002): 1266-1272.
- Adinolfi LE, Gambardella M, Andreana A, et al. Steatosis accelerates the progression of liver damage of chronic hepatitis C patients and correlates with specific HCV genotype and visceral obesity. Hepatology 33 (2001): 1358-1364.
- Alter HJ, Seeff LB. Recovery, persistence, and sequelae in hepatitis C virus infection: a perspective on long-term outcome. Semin Liver Dis 20 (2000): 17-35.
- Fabrizi F, Dixit V, Martin P, et al. Hepatitis C virus infection and kidney disease: a meta-analysis. Clin J Am Soc Nephrol 7 (2012): 549-557.
- Dimova RB, Zeremski M, Jacobson IM, et al. Determinants of hepatitis C virus treatment completion and efficacy in drug users assessed by meta-analysis. Clin Infect Dis 56 (2013): 806-816.
- Armstrong GL, Wasley A, Simard EP, et al. The prevalence of hepatitis C virus infection in the United States, 1999 through 2002. Ann Intern Med 144 (2006): 705-714.
- Platt L, Easterbrook P, Gower E, et al. Prevalence and burden of HCV co-infection in people living with HIV: a global systematic review and meta-analysis. Lancet Infect Dis 16 (2016): 797-808.
- Sharma S, Carballo M, Feld JJ, Janssen HL. Immigration and viral hepatitis. J Hepatol 63 (2015): 515-522.
- Gower E, Estes C, Blach S, Razavi-Shearer K, Razavi H. Global epidemiology and genotype distribution of the hepatitis C virus infection. J Hepatol 61 (2014): S45-57.


 Impact Factor: * 6.2
Impact Factor: * 6.2 Acceptance Rate: 76.33%
Acceptance Rate: 76.33%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 