Insilco Peptide Design: A Novel Step Forward to Combat Β-Lactamase Mediated Antimicrobial Resistance
Varshaa Arer, Deepesh Nagarajan, Debasish Kar*
Department of Biotechnology, Ramaiah University of Applied Sciences, Bangalore, India
*Corresponding author: Debasish Kar, Department of Biotechnology, Ramaiah University of Applied Sciences, Bangalore, India.
Received: 01 December 2024; Accepted: 09 December 2024; Published: 07 January 2025
Article Information
Citation: Varshaa Arer, Deepesh Nagarajan, Debasish Kar. Insilco Peptide Design: A Novel Step Forward to Combat Β-Lactamase Mediated Antimicrobial Resistance. Fortune Journal of Health Sciences. 7 (2024): 14-25.
View / Download Pdf Share at FacebookAbstract
Antimicrobial resistance (AMR) is one of the most important global health challenges, driven by the ability of bacteria to produce β-lactamase enzymes that hydrolyze β-lactam antibiotics and, therefore, neutralize their effects. This poses a challenge in inhibitor development because the classes A, B, C, and D of β-lactamases show distinct catalytic mechanisms. In this study, in silico approaches involving protein modelling of 20 proteins, and molecular docking have been used to design pentapeptides inhibitors. The study involves specific β-lactamase variants, such as CAE-1 and PER-1 of Class A, VIM-24 and IMP-26 of Class B, CMY-37 and FOX-1 of Class C, and OXA-372 and OXA-50 of Class D β-lactamases. Modelled three-dimensional enzyme structures facilitated the design of peptides against the active site, discovery of high-affinity interactions, and inhibition of enzymatic activity. These results outline the potential of peptide-based therapeutics in overcoming resistance caused by β-lactamase, thereby paving ways to innovative treatments against drug-resistant bacterial infections and ultimately towards improving global public health.
Keywords
<p>β-lactamase, pentapeptides, docking, modelling, antimicrobial-resistance</p>
Article Details
Introduction
β-lactamases are bacterial enzymes that make the bacterium resistant to the β-lactam drugs by degrading the β-lactam ring further; it, therefore, loses its effectiveness. β-lactamases are enzymes that split the β-lactam ring of penicillins, cephalosporins, monobactams, and carbapenems, and with that, antibiotics turn out to be inanimate against bacteria that have developed resistance towards them [1]. Some other clinically relevant microorganisms, among which are Escherichia coli and Klebsiella pneumoniae, have well been shown to produce an enormous amount of class A β-lactamases falling into one of the highest classes of this enzyme class. Maintaining the effectiveness of the β-lactam antibiotics remains to be the need of breaking down the resistance provided by β-lactamases [2]. This classification is based upon their amino acid sequences and structural homologies; hence, the four primary classes of them are (A, B, C, and D). The Class A β-lactamases, usually called serine β-lactamases, are also of much interest since it is the most widely dispersed and significantly important in medicinal use. These enzymes contain an active serine residue that helps in the formation of a covalent acyl-enzyme intermediate to achieve the breaking of the β-lactam ring [3]. The pentapeptides design can be highly potent for building new useful drugs, especially from the area of antibiotic-resistant drugs.
Pentapeptides are five-amino acid residues, offering a number of advantages over the inhibitor based on a larger protein. They are compact in size and straight in shape, which enhances cell permeability, making it easy to penetrate bacterial cells to target intracellular components like β-lactamases [4]. Though pentapeptides are small, computer-assisted methods can design them to be strong in the sense that they can target particular enzymes or proteins by augmenting the binding interactions and structural compatibility [5]. More importantly, linear pentapeptides have less chance to induce an immune response as compared to the larger protein inhibitors. That leads to reduced chances of having harmful effects and increases its use prospects as drugs [4]. The strategy involves synthesis of linear pentapeptides inhibitors combined with approaches for in silico designs and may hold some bright outlook to the vanishing antibiotic resistance and the regain activity of the existing ones over the resistant strains of the bacteria [6]. In silico methods allow predictions of possible inhibitors and enzymes interaction, their binding affinities determination, and structural complementarity as well. Researchers can effectively design inhibitors that bind and react with specific enzymes by studying the three-dimensional structures and inherent properties of target β-lactamases [3]. This research study deals with computational approaches that attempt to produce a pentapeptides with specificity towards the active site of all classes of β-lactamase enzymes. The approach makes accurate modeling and simulation possible, aiding in the discovery of possible pentapeptides sequences that might attach and inhibit the enzymatic activity of β-lactamases. Using highly competent molecular modeling as well as docking techniques, this piece of research work gathers critical understanding over the bonding dynamics of the pentapeptides with the enzyme involved. The discoveries are optimistic and hopeful for the invention of novel inhibitors to vanquish antibiotic-resistant bacteria populations.
Methodology
Homology modeling and Sequence analysis
Clustal Omega is used for multiple sequence alignments to check similarity among the proteins. By choosing homologous sequences, protein templates were selected as described in the Table 1 from the following bacterial species. Using MODELLER 10.2 for homology modeling, best energy models were selected and model was energy minimized using swiss pdb viewer for further inspection [7, 8].
Table 1: Overview of templates used for β-lactamase
|
Query Protein |
Selected Template |
Percentage Similarity (%) |
Template ID |
Resolution (Å) |
|
Class A β-lactamase |
||||
|
BCK-1 β-lactamase of Klebsiella pneumoniae |
Ancestral PNCA synthetic construct |
61.07 |
4C6Y |
1.8 |
|
PER-1 β-lactamase of Providencia rettgeri |
PER-1 of Psuedomonas aeruginosa |
87.95 |
1E 25 |
1.9 |
|
CAE-1 β-lactamase of Comamonas aquatica |
Ancestral PNCA synthetic construct |
58.1 |
4C6Y |
1.8 |
|
TLA-1 of Klebsie β-lactamase lla michiganensis |
TLA-3 of Serratia marcescens |
99.64 |
5GS8 |
1.59 |
|
GES-22 β-lactamase of Acinetobacter baumannii |
GES-11 of Acinetobacter baumannii |
99.65 |
3V3R |
1.9 |
|
Class B β-lactamase |
||||
|
IMP-26 β-lactamase of Enterobacter cloacae |
IMP-26 β-lactamase of Escherichia coli |
95.53 |
7XHW |
1.7 |
|
IND-2 β-lactamase of Chryeobacterium indologens |
IND-2 β-lactamase from Escherichia coli |
99.59 |
3L6N |
1.65 |
|
SPM-1 β-lactamase of Acinetobacter baumannii |
IND-2 β-lactamase from Psuedomonas aeruginosa |
99.59 |
2FHX |
1.9 |
|
TMB-1 β-lactamase of Enterobacter hormaechei |
IND-2 β-lactamase from Psuedomonas aeruginosa |
67.26 |
4WD6 |
2.2 |
|
VIM-24 β-lactamase of Psuedomonas aeruginosa |
VIM-24 β-lactamase of Acientobacter baumannii |
99.59 |
5YD7 |
1.7 |
|
Class C β-lactamase |
||||
|
CMY-4 β-lactamase of Escherichia coli |
CMY-136 β-lactamase of Escherichia coli |
99.45 |
6G9T |
1.6 |
|
CMY-37 β-lactamase of Citrobacter freundii |
CMY-136 β-lactamase of Escherichia coli |
95.26 |
6G9T |
1.6 |
|
FOX-1 β-lactamase of Kelbsiella pnuemoniae |
FOX-4 β-lactamase of Escherichia coli |
96.4 |
5CGS |
1.63 |
|
ACT-1 β-lactamase of Enterobacter mori |
AmpC β-lactamase of Enterobacter cloacae |
85.04 |
6LC7 |
1.4 |
|
AmpC β-lactamase of Erwinia rhapontici |
AmpC β-lactamase of Escherichia coli |
66.99 |
5GGW |
1.76 |
|
Class D β-lactamase |
||||
|
OXA-4 β-lactamase of Psuedomonas aeruginosa |
OXA-1 β-lactamase of Escherichia coli |
99.8 |
4MLL |
1.34 |
|
OXA-2 β-lactamase of Psuedomonas aeruginosa |
OXA-2 β-lactamase of Salmonella enterica |
99.61 |
1K38 |
1.5 |
|
OXA-204 β-lactamase of Klebsiella pnuemoniae |
OXA-48 β-lactamase of Klebsiella pnuemoniae |
98.87 |
6PXX |
1.5 |
|
OXA-50 β-lactamase of Psuedomonas aeruginosa |
OXA-23 β-lactamase of Acinetobacter baumannii |
44.35 |
4JF6 |
2.5 |
|
OXA-372 β-lactamase of Citrobacter freundii |
OXA-48 β-lactamase of Klebsiella pnuemoniae |
42.68 |
5FAQ |
1.96 |
Model evaluation and Analysis
The quality of the model was checked by the use of ProQ, ModFOLD, PROCHECK, and Verify 3D. CASTp measured the volume of the groove of the active site. Secondary structure prediction has been done by PREDICT PROTEIN and PSIPRED. Structural alignment was done by RMSD, and contact potentials are analyzed in PyMOL [9-19].
Peptide model preparation
We created a library of 1000 randomly selected PDB entries from the PDB database as of November, 25th 2023 (Text S1). We extracted unique protein chains for every PDB entry for downstream processing. For every unique protein chain, we used DSSP [20] to annotate the secondary structures. DSSP also assigns backbone phi and psi dihedral angles for every protein residue. We compiled a database of phi and psi angles for every residue found in the loop region of every unique protein chain. This database possessed 282,002 unique phi-psi pairs (Text S2), making it a representative subset of the loop backbone conformational space. A Ramchandaran plot (phi-psi map) for this data is provided in (Supplementary Figure S1). We found a greater prevalence of residues adopting dihedral angles in the positive phi space (0 to 180°) compared to the generalized Ramachandran plot for all proteins.
We then constructed a library of 1000 polyalanyl pentapeptides (AAAAA) using backbone dihedral angles randomly selected from our database of phi-psi pairs (Dataset S1). These pentapeptides created using a representative sample of phi-psi angles provided a large sampling of peptide conformational space, which otherwise would not have been possible as out downstream methods treat ligands as rigid bodies. These 1000 polyalanyl pentapeptides were then site specifically docked using Autodock Vina [21] against a 20-member protein dataset respresenting class A-D β lactamases.
Peptide sequence design
For every protein-polyalanyl peptide pair successfully docked usind Autodock Vina during step 4, we performed sequence design on the polyalanyl peptides using Rosetta [22, 23]. Rosetta fixed backbone design (fixbb.static.linuxgccrelease, Rosetta 3.14) computationally mutates every alanine to the most appropriate residue required to minimize the global rosetta energy function. During this step the backbone dihedral angles are not altered. Subsequently we performed all-atom relaxation (relax.static.linuxgccrelease, Rosetta 3.14) for both the designed pentapeptide and target β lactamase. All-atom relaxation perturbs backbone and sidechain dihedral angles without introducing new mutations. This step is required in order to find the global energy minima for newly designed pentapaptides. For every β lactamase class (4 classes), we selected and reported the top 25 pentapeptides displaying the lowest global rosetta energy minima, indicating the strongest predicted binding. Structures for every designed protein-peptide interaction are provided in Dataset S2. The approach employed in current study is summarized in Figure 1 that in itself depicts the sequential steps followed during this research.
Results and discussion
Homology modelling
Homology modelling of 20 various proteins from classes A, B, C and D β-lactamases were analyzed using molpdf (molecular probability density function), DOPE score, and GA341 scores. The molpdf values determine how well the models fit the input constraints, with lower scores reflecting better fitting. Lower DOPE scores, a statistical energy-based measure of protein model quality, suggest more favorable energetic landscape. The GA341 score on a scale from 0 to 1, indicates the reliability of the model based on which very close values to 1 indicate an outstanding level of dependability (John, 2003). All these models have obtained a GA341 score of 1.00000 (for the whole set), thus proving its reliability in this study. Out of the truncated and modeled proteins alone, some models were preferentially better based on QMEAN Z-scores. One model in each gene were selected according to the lowest DOPE score. The predicted structures were further refined by post-modelling energy minimization, resulting in the selection of stable models that are appropriate to be used for subsequent analyses.
Model Validation
Homology modelling of 20 β-lactamase proteins (5 in each class A, B, C and D) was performed as described above and the energy minimisation predicted structures were submitted to an array of stringent structural assessment tools to validate their accuracy and reliability for subsequent studies. The global and local quality of the generated models were assessed using various tools including PROQ (LGscore/MaxSub), ModFOLD, Procheck, VERIFY3D, ERRAT, ProSA and PSIPRED [9-19]. PROQ scores validated structural quality metrics, and a high LG score suggested confidence in model accuracy. The reliability scores from ModFOLD were consistent with expected structural landscape landmarks, confirming the robustness of our models. Procheck performed Ramachandran plots with the majority of the residues in the most favorable and additionally allowed regions, suggesting good stereochemistry.
All other models displayed comparable 3D-1D profiles, with >90% of residues having compatible profiles from VERIFY3D analysis supporting their structural compatibility. The average ERRAT scores (85-95%) exhibited very few gross structural errors overall. ProSA scores afforded extremely high Z-scores typical for high-resolution experimental structures in the Protein Data Bank, while PSIPRED secondary structure predictions showed excellent agreement with expected motifs, further confirming model stability and integrity. Altogether, that confirmed the quality of all generated models suitable for upstream applications: docking studies, molecular dynamics simulations or inhibitor designing. Table 2 summarizes the results of validation, and Ramachandran plots for each model. These results demonstrate the power of the modelling approach and serve as a subsequent basis for further computational and experimental studies.
Table 2: Summary of model validation of β-lactamases
|
TOOL |
Class A β-lactamase |
|||||
|
|
BCK-1 |
PER-1 |
CAE-1 |
TLA-1 |
GES-22 |
|
|
PRO Q LG score/Max Sub |
7.43/-0.574 |
8.741/-0.507 |
8.570/-0.670 |
10.499/-0.713 |
8.025/-0.574 |
|
|
ModFOLD |
2.469/0.6278 |
4.59/0.7034 |
1.018/0.6676 |
3.995/0.7096 |
2.539/0.730 |
|
|
Procheck |
87.40% |
93.3 |
90.60% |
91.5 |
94.4 |
|
|
VERIFY3D |
47.92 |
63.49 |
65.12 |
82.52 |
79.44 |
|
|
ERRAT |
77.174 |
92.25 |
83.0258 |
96.3768 |
89.5911 |
|
|
ProSA |
-5.69 |
-6.63 |
-6.33 |
-7.86 |
-6.5 |
|
|
Class B β-lactamase |
||||||
|
IMP-26 |
IND-2 |
SPM-1 |
TMB-1 |
VIM-24 |
||
|
PRO Q LG score/Max Sub |
8.017/-0.536 |
7.391/-0.537 |
7.619/-0.556 |
7.567/-0.593 |
9.607/-0.666 |
|
|
ModFOLD |
3.361/0.7174 |
5.561/0.6948 |
4.329/0.7060 |
4.314/0.7062 |
7.497/0.6813 |
|
|
Procheck |
91.40% |
89 |
91.2 |
93.8 |
94.3 |
|
|
VERIFY3D |
83.74 |
86.83 |
80.53 |
89.8 |
79.7 |
|
|
ERRAT |
81.9742 |
84.44 |
88.477 |
82.969 |
83.1897 |
|
|
ProSA |
-7.58 |
-7.54 |
-8.17 |
-7.61 |
-7.66 |
|
|
Class C β-lactamase |
||||||
|
CMY-4 |
CMY-37 |
FOX-1 |
ACT-1 |
AmpC |
||
|
PRO Q LG score/Max Sub |
7.131/-0.536 |
8.169 / -0.427 |
10.055 / -0.498 |
7.728 /-0.378 |
6.507/-0.320 |
|
|
ModFOLD |
3.67/0.7134 |
7.314/2.463 |
0.7320/2.43 |
0.7105/3.919 |
0.7197/3.193 |
|
|
Procheck |
94.20% |
93.60% |
93.50% |
93.60% |
93.20% |
|
|
VERIFY3D |
81.63% |
80.58% |
87.43% |
83.99% |
85.49% |
|
|
ERRAT |
85.6749 |
88.5475 |
80.5085 |
79.6089 |
87.8873 |
|
|
ProSA |
-8.55 |
-8.92 |
-8.61 |
-8.64 |
-9.24 |
|
|
Class D β-lactamase |
||||||
|
OXA-4 |
OXA-2 |
OXA-204 |
OXA-50 |
OXA-372 |
||
|
PRO Q LG score/Max Sub |
9.435/-0.755 |
7.504/-0.574 |
8.681/-0.692 |
7.002/-0.583 |
6.303/-0.556 |
|
|
ModFOLD |
7.685/0.6802 |
5.109/0.6986 |
3.112/0.7208 |
6.68/0.6865 |
2.545/0.729 |
|
|
Procheck |
89.3 |
90.03 |
90.9 |
88.7 |
90 |
|
|
VERIFY3D |
69.2 |
77.82 |
86.64 |
91.98 |
70.04 |
|
|
ERRAT |
93.9759 |
89.068 |
93.305 |
74.1935 |
80.41 |
|
|
ProSA |
-7.84 |
-6.86 |
-8.09 |
-7.41 |
-7.03 |
|
Note: Pro (LG):>1.5 fairly good; >2.5 very good; >4 extremely good. ProQ (MX): >0.1 fairly good; >0.5 very good; >0.8 extremely good. ModFOLD (Q): >0.5 medium confidence; >0.75 high confidence. ModFOLD (P): <0.05 medium confidence; <0.01 high confidence.PROCHECK: A good quality smodel would be expected to have over 90% residues in the most favored region of Ramachandran plot. Verify_3D: It is the measurement of compatibility of the 3D structure of the model generated to its amino acid sequence on a % scale of 100. ERRAT: Good high-resolution structures generally produce values around 95% or higher. For lower resolutions the average overall quality factor is around 91%. ProSA: more negative better the quality.
Auto-dock and peptide designing of β-lactamases
Class A β-lactamases
The AutoDock and Rosetta scores given in the table reveal the inhibition ability from examining top 25 of peptides that were designed against class A β-lactamases. Peptide plot — analysis of the results produced by Rosetta, providing information on docking scores and sequences of the peptides; this outputs focus mainly on five top scoring peptides identified by their Rosetta scores (Figure 2). All these peptides show important docking scores, and the top five have very low (more negative) values (Table 3), demonstrating a strong ability to bind. Further evidence for the likely stability and efficacy of these peptides to be able to bind their target proteins comes from Rosetta scores.
Table 3: Detailed scoring and functional composition of top 25 peptides in class A β-lactamases
|
Class A β-lactamases |
Autodock Scores |
Rosetta Scores |
Sequence |
|
Peptide 0962 |
-7.247 |
-895.731 |
GGKST |
|
Peptide 1333 |
-7.393 |
-890.636 |
AGGST |
|
Peptide 0354 |
-7.094 |
-886.526 |
AGGST |
|
Peptide 1177 |
-7.194 |
-886.336 |
AGMSV |
|
Peptide 1515 |
-7.201 |
-885.857 |
GGKTT |
|
Peptide 1585 |
-7.091 |
-660.41 |
AGLTY |
|
Peptide 1769 |
-7.423 |
-656.098 |
AEGSS |
|
Peptide 1144 |
-7.324 |
-654.056 |
AGKTY |
|
Peptide 0299 |
-7.154 |
-652.067 |
AAEGT |
|
Peptide 1513 |
-7.126 |
-649.613 |
AAETT |
|
Peptide 1791 |
-7.23 |
-648.181 |
AAEKT |
|
Peptide 1959 |
-7.427 |
-648.031 |
AAETT |
|
Peptide 1172 |
-7.066 |
-645.99 |
AAETT |
|
Peptide 1898 |
-7.203 |
-644.026 |
AASST |
|
Peptide 1305 |
-7.349 |
-642.12 |
AADSS |
|
Peptide 0654 |
-7.378 |
-640.283 |
AAEST |
|
Peptide 1647 |
-7.122 |
-639.08 |
AADIS |
|
Peptide 1325 |
-7.238 |
-635.953 |
AAISS |
|
Peptide 0158 |
-7.274 |
-635.544 |
AADSS |
|
Peptide 1107 |
-7.109 |
-635.188 |
AASST |
|
Peptide 0055 |
-7.145 |
-627.485 |
ADQGT |
|
Peptide 0397 |
-7.325 |
-626.058 |
ADQGS |
|
Peptide 1277 |
-7.139 |
-624.273 |
ANEGG |
|
Peptide 1274 |
-7.019 |
-623.291 |
AGKTY |
|
Peptide 0075 |
-7.127 |
-619.279 |
AAEKT |
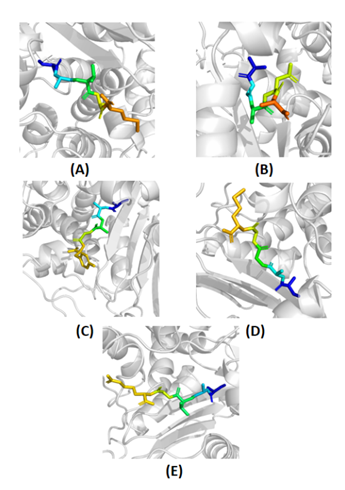
Figure 2: Binding conformations of top five peptide inhibitors of class A β-lactamases. Visualization of top five peptides inhibitors (A-E) in the active site of class A β-lactamases. (A) Peptide 0962 (B) Peptide 1333 (C) Peptide 0354 (D) Peptide 1177 (E) Peptide 1515. The peptides are depicted in stick representation emphasizing their spatial arrangements and binding poses
Peptide - 1333: AGGST; AutoDock score −7.393; Rosetta score −890.636 Ala and Gly are used to promote hydrophobic interactions, whereas Ser and Thr provide polar characteristics which can create potential hydrogen bonds. Such integration through binding means being comprehensive and equitable [20]. Peptide 0962 (sequence: GGLST) has an AutoDock score of -7.247 and a Rosetta score of -895.731, respectively. Flexible multiple glycine residues (Gly) and charged lysine (Lys) that provide a cation for electrostatic interaction with negatively charged targets [21]. The polar side chains of serine and threonine increase hydrogen bond capacity. Despite having the sequence AGGST, similar to peptide 1333, Peptide 0354 had an AutoDock score of −7.094 and a Rosetta score of −886.526. Alanine and glycine increase stability and flexibility, while serine and threonine promote hydrogen bonding [22]. Peptide 1177 (AGMSV) involves hydrophobic interaction by methionine (Met) and valine (Val), potential hydrogen bonding interactions by alanine and serine, giving it AutoDock score of -7.194 and a Rosetta Score of -886.336). Similar patterns are observed in the sequences of top five peptides such as a high frequency for glycine which provide flexibility and Alanine also proving its position for hydrophobic stability. The proportion of hydrophobic (e.g., A, M, V) and polar residues (e.g., S, T) increases the binding abilities in a variety of environments [23]. These features indicate a systematic design strategy that combines adaptability, robustness, and specificity for binding. The presence of both hydrophobic and polar residues enables flexibility under different conditions, further enhancing the therapeutic relevance of the peptides. Recent studies have reported similar insights on peptide design principles and noted that balance in amino acid composition supports the binding affinity and specificity [24].
Class B β-lactamases
The peptides generated through Rosetta are evaluated with Docking scores and sequences that can inform on their ability to bind a known protein of interest (Table 4). The top five peptides show a bound docking score with quite low (more negative) values, which illustrates higher binding affinities of the peptides (Figure 3). These findings are consistent with recent reports that property-based docking metrics can serve as better predictor of peptide-protein interactions [25]. These Rosetta scores provides further proof of concept for the stability and functionality of these peptides to bind their respective target proteins.
Table 4: Detailed scoring and functional composition of top 25 peptides in class B β-lactamases
|
Class B β-lactamases |
Autodock Scores |
Rosetta Scores |
Sequence |
|
Peptide 1166 |
-7.081 |
-734.626 |
AGKMS |
|
Peptide 0058 |
-7.062 |
-725.719 |
AAAAT |
|
Peptide 0270 |
-6.904 |
-724.558 |
AAAAT |
|
Peptide 1640 |
-6.995 |
-724.476 |
NGLST |
|
Peptide 1789 |
-6.866 |
-723.684 |
AAGKM |
|
Peptide 1051 |
-7.109 |
-723.512 |
AAGGL |
|
Peptide 1379 |
-7.156 |
-723.267 |
AAAGI |
|
Peptide 1778 |
-6.897 |
-721.488 |
AAQGK |
|
Peptide 1300 |
-6.955 |
-717.958 |
AQGKS |
|
Peptide 1150 |
-7.097 |
-717.039 |
AAGGY |
|
Peptide 0690 |
-6.922 |
-715.934 |
AAAGI |
|
Peptide 0787 |
-7.007 |
-715.66 |
AAAGI |
|
Peptide 0720 |
-7.134 |
-715.186 |
AAAAT |
|
Peptide 0994 |
-7.112 |
-714.352 |
AAAGI |
|
Peptide 0984 |
-6.888 |
-713.614 |
ANNGK |
|
Peptide 0453 |
-7.065 |
-712.725 |
AAAAT |
|
Peptide 0592 |
-7.076 |
-711.265 |
NNGKW |
|
Peptide 0801 |
-7.158 |
-710.791 |
AAAGI |
|
Peptide 0998 |
-6.973 |
-710.036 |
DGKMS |
|
Peptide 0755 |
-6.968 |
-709.997 |
AAAST |
|
Peptide 0332 |
-7.141 |
-708.732 |
NGLST |
|
Peptide 1225 |
-7.151 |
-708.068 |
NNGMS |
|
Peptide 1465 |
-7.248 |
-707.534 |
AEGYV |
|
Peptide 1179 |
-7.016 |
-702.789 |
NNGKM |
|
Peptide 1817 |
-7.064 |
-701.623 |
NGKKM |
Figure 3: Binding conformations of top five peptide inhibitors of class B β-lactamases. Visualization of top five peptides inhibitors (A-E) in the active site of class B β-lactamases. (A) Peptide 1465 (B) Peptide 0058 (C) Peptide 0270 (D) Peptide 1640 (E) Peptide 1789. The peptides are depicted in stick representation emphasizing their spatial arrangements and binding poses
Peptide 1465 (Autodock score: -7.248, Rosetta score: -707.534; AEGYV). In this peptide, glutamic acid (Glu) provides a negative charge to enhance electrostatic contacts, tyrosine (Tyr) provide an aromatic property for π-π stacking interactions, and valine (Val) has hydrophobic property for enhancing the binding stability due to hydrophobic interactions [24]. The second peptide 1166 (AGKMS) with Autodock score −7.081 and Rosetta score −734.626 It has Lys (positive) as an amino acid, which could form salt bridges with negative targets. Furthermore, methionine (Met) adds hydrophobicity and serine (Ser) participates in hydrogen bonding, thereby providing a delicate balance of interactions [26].Peptide 0058 (AAAAT) has an Autodock score of−7.062 and Rosetta Score of −725.719 The high frequency of alanine (Ala) allows for hydrophobic interactions, which stabilize the structure. In contrast, threonine (Thr) offers a polar hydroxyl group that can participate in potential hydrogen bonding [25]. Such feature might enhance the association with intended proteins. Peptide 0270: AAAAT autodock score -6.904 Rosetta score − 724.558 the relevance of alanine for stability is illustrated with this sequence comparison using peptide 0058 as a model. The introduction of threonine in the sequence is also said to be increase chances for hydrogen bonding and hence may enhance the binding affinities. Peptide1789: AAGKM, Autodock score: -6.866, Rosetta score:-723.684 It consists of Glycine (Gly) which gives flexibility, lysine (Lys) that provides positive charge together with methionine (Met), to confer hydrophobicity, thereby enhancing the ability of peptide to achieve hydrophobic interactions.
Across the top five peptides, there are a number of commonalities. Peptides 0058, 0270 and 0720 contained lists of four contiguous Ala which indicate a marked focus on hydrophobic contacts. Peptide 1465 contains glutamic acid (Glu), which carries a negative charge, allowing for electrostatic interactions. In contrast, Peptide 1166 features lysine (Lys), a positively charged residue capable of electrostatic interaction with negatively charged targets. The amino acid tyrosine (Tyr), which possesses the ability to π-stack, is present in peptide 1465. The sequences often contain a combination of hydrophobic amino acids (e.g., Ala, Val, Met) accompanied by polar ones (e.g., Ser, Thr), consistent with an evolutionary strategy that balances binding affinity and flexible interactions in various contexts. Peptides 1166 and 1789 also contain glycine (Gly), which enhances flexibility, while the high mole fraction of alanine provides hydrophobic stability. Crossover of these traits enables binding under diverse contexts [27]. The frequent occurrences of alanine and glycine residues in these peptides probably reflect a particular focus on structural stability and flexibility in extracellular locations. It could also also increase hydrophobic interactions if alanine is repeated or permit structural flexibility with glycine. Charged and polar residues based on various amino acids, for example, lysine, serine, and threonine — suggesting the potential to create hydrogen bonds and electrostatic interactions with target proteins [25].The rich composition of amino acids including residue types that are hydrophobic, polar and charged ensures that the peptides can possibly interact with a range of biological targets. It is reinforced with repeating alanine residues giving it strength and glycine residues allowing for flexibility. The mixture allows the peptides to adopt favorable binding conformations [24]. These patterns reflect the state of knowledge in peptide design, whereby an optimal profile of amino acid features is often sought to improve binding affinity and specificity. Repeated occurrence of some specific residues and insertion of charged and polar amino acids on these peptides suggest that they were intentionally made to enhance their ability to interact with target proteins.
Class C β-lactamases
Analysis of the peptides found for Rosetta provides important insights related to docking scores and sequences with a detailed discussion focused on the top five peptides (Table 5). The docking scores for these peptides are high and top 5 have lowest (more negative) values suggestive of higher binding affinities (Figure 4). In addition, the Rosetta scores are consistent with potential stability and viability of these peptides in their ability to bind the target proteins.
Table 5: Detailed scoring and functional composition of top 25 peptides in class B β-lactamases
|
Class C β-lactamases |
Autodock Scores |
Rosetta Scores |
Sequence |
|
Peptide 1183 |
-8.483 |
-1133.67 |
AGGGS |
|
Peptide 0609 |
-8.349 |
-1129.12 |
AAAGS |
|
Peptide 1562 |
-8.351 |
-1128.58 |
AAAAS |
|
Peptide 0038 |
-8.664 |
-1126.19 |
AAGGG |
|
Peptide 0421 |
-8.347 |
-1121.99 |
ADGST |
|
Peptide 0374 |
-8.594 |
-1121.12 |
AGGMS |
|
Peptide 0027 |
-8.362 |
-1120.24 |
AGSST |
|
Peptide 0277 |
-8.464 |
-1120.24 |
AAAAG |
|
Peptide 0543 |
-8.397 |
-1119.54 |
ADGGG |
|
Peptide 0685 |
-8.596 |
-1118.74 |
AGGSS |
|
Peptide 1122 |
-8.375 |
-1118.07 |
AAGSS |
|
Peptide 0926 |
-8.615 |
-1115.82 |
AAGGV |
|
Peptide 1129 |
-8.625 |
-1114.69 |
AAGSV |
|
Peptide 1854 |
-8.329 |
-1114.07 |
AAAGS |
|
Peptide 0285 |
-8.7 |
-1114.02 |
AGGGM |
|
Peptide 0720 |
-8.39 |
-1112.93 |
AAMST |
|
Peptide 0767 |
-8.524 |
-1111.99 |
ARGST |
|
Peptide 1091 |
-8.463 |
-1109.92 |
AAANG |
|
Peptide 1174 |
-8.372 |
-1109.46 |
AAGST |
|
Peptide 0653 |
-8.445 |
-1109.18 |
AAGST |
|
Peptide 0994 |
-8.478 |
-1105.49 |
AAGSY |
|
Peptide 1952 |
-8.479 |
-1100.92 |
AGSST |
|
Peptide 0965 |
-8.577 |
-1100.58 |
AAAAG |
|
Peptide 1974 |
-8.626 |
-1099.67 |
AAGST |
|
Peptide 1139 |
-8.459 |
-1092.9 |
ARGGS |
Figure 4: Binding conformations of top five peptide inhibitors of class C β-lactamases. Visualization of top five peptides inhibitors (A-E) in the active site of class C β-lactamases. (A) Peptide 0038 (B) Peptide 0285 (C) Peptide 1974 (D) Peptide 1183 (E) Peptide 0609. The peptides are depicted in stick representation emphasizing their spatial arrangements and binding poses
Peptide 0038: Sequence: AAGGG (Autodock score −8.664, Rosetta score −1126.19). This peptide contains resolutions A, which increase hydrophobic interactions, and G, which enhance flexibility. The repeated occurrence of glycine enhances its flexibility to accommodate various conformations, and promotes high-binding affinity. Researchers have observed comparable results regarding the role of glycine in regards to structural flexibility on a similar timescale. Studies show peptide-binding [28]. Peptide 0285 with the sequence AGGGM, Autodock Score (-8.7), Rosetta score (-1114.02). This repeats contains glycine which increases the flexibility of the molecule and methionine (Met) which increases its hydrophobic nature. These residues allow the peptide to engage in hydrogen interactions but should retain flexible structure. Data from recent study support the hypothesis that methionine plays any essential role by stabilizing hydrophobic cores [26]. Peptide 1974 is a peptide with sequence AAGST and Autodock score -8.626/Rosetta: -1099.67. This is a polar sequence, which contains alanine (Ala) for stabilization and serine (Ser) and threonine (Thr). Serine and threonine have hydroxyl groups that can form hydrogen bonds. Such property would likely ameliorate the interaction of these amino acids with target proteins. A study point out similar contributions to binding affinity from hydroxyl-bearing residues [29]. Peptide 1183, an Autodock score of -8.483 and Rosetta score -1133.67 peptide with AGGGS sequence. This series has a high preference for glycine for its facilitatory role in flexibility, and includes serine, which is capable of hydrogen bonding. Several glycine residues suggest an emphasis on flexibility, while serine could aid in the specificity of binding. In some of the most recent computational studies to directly address this question, researchers have shown that glycine's propensity for adaptability in peptide backbone conformation is predictive of local index conformational propensities41—42 and thereby able to guide and even improve peptide design [30]. Peptide 0609 the sequence is AAAGS, and it has an Autodock score of -8.349 and a Rosetta score of -1129.12. Here we adapted the peptide with alanine to increase the stability, and glycine for increased flexibility, and serine where hydrogen bonds may form. The balance of these residues reflects a broader binding interaction strategy. Previous studies have supported the hydrophobic contribution to peptide stability of alanine [24].
There are trends among the top five peptides. This shows a particular focus on hydrophobic interactions, as peptides 0058, 0270, and 0720 each contain four consecutive alanine (Ala) residues in their sequence. This point is important since there are both polar and charged residues. Peptide 1465, for example, has glutamic acid (Glu) in it which adds a negative charge and can facilitate electrostatic interactions. In contrast, in peptide 1166 there is lysine (Lys), a positively charged residue that can bind to negatively charged targets. The splenocyte-derived amino acid tyrosine (Tyr) of peptide 1465 has the potential to π-stack. The sequences often include hydrophobic amino acids (Ala, Val, Met) in addition to polar amino acids (Ser, Thr), which suggests a balanced approach between maximizing binding potential and facilitating flexible interactions in various environments. Peptides 1166 and 1789 also contain glycine (Gly), which increases flexibility, as well as an abundance of alanine that is indicative of hydrophobic stability. This combination of properties yields successful binding in diverse context. The patterns suggest that the top 5 peptides are actually engineered to do this considering the best interactions with target proteins. More specifically, the high prevalence of alanine enhances hydrophobic interactions, while the presence of charged and polar residues fosters hydrogen bonding and electrostatic interactions. The combination of those features provides adaptability to bind in various conditions [31]. Incorporating glycine into the peptides increases their flexibility so that they can change shape to fit different targets better. Collectively, these data illustrate a selective design protocol for peptides and underscore the importance of sequence composition in controlling stability, affinity, and specificity of binding.
Class D β-lactamases
Docking scores and sequences for the investigated Class D β-lactamases peptides derived from Rosetta are provided in Table 6, with the five highest scoring (most favorable binding profiles) of each peptide listed in Figure 6. These peptides have high binding potential because their amino acid compositions help to maintain stability, flexibility and the possibility of interaction [32]. Peptide 1139 (AGGGS): Autodock score= −7.495, Rosetta score = -770.94; contains glycine and serine residues that contribute to flexibility and promote hydrogen bond formation for protein-specific interactions [33]. Peptide 1856 (ADGLT): Autodock score = −7.549, Rosetta score = −765.918 with alanine for stability, glycine for flexibility and threonine and aspartic acid contributing polar interactions via hydrogen bonding promoting binding efficiency [34]. Peptide 1642 (ARDGT): Autodock score = -7.487, Rosetta score =-763.023 Features: Arginine for salt bridge interaction, Glycine allows flexibility of the peptide backbone and Threonine also helps in additional polar interactions. Peptide 1520 (GGGLS): Autodock score = −7.47, Rosetta score = −762.557; bias towards glycine residues to promote mobility, serine residues to ensure high numbers of hydrogen bonding interactions for strong stable binding [35]. Peptide1065 (EGGGS): Autodock score = -7.606, Rosetta-friendly score = -760.441; glutamate serves to ionic interactions and glycine ensures flexibility purposes, serine provides specificity with two hydrogen bonds [36]. These results highlight the importance of integrating hydrophobicity, polar nature, and structural flexibility for optimal peptide recognition by Class D β-lactamases.
Table 6: Detailed scoring and functional composition of top 25 peptides in class B β-lactamases
|
Class D β-lactamases |
Autodock Scores |
Rosetta Scores |
Sequence |
|
Peptide 1139 |
-7.495 |
-770.94 |
AGGGS |
|
Peptide 1856 |
-7.549 |
-765.918 |
ADGLT |
|
Peptide 1642 |
-7.487 |
-763.023 |
ARDGT |
|
Peptide 1520 |
-7.47 |
-762.557 |
GGGLS |
|
Peptide 1065 |
-7.606 |
-760.441 |
EGGGS |
|
Peptide 1241 |
-7.591 |
-758.836 |
GGGLS |
|
Peptide 1637 |
-7.633 |
-758.449 |
EGGGS |
|
Peptide 0320 |
-7.477 |
-758.232 |
NGGGV |
|
Peptide 1305 |
-7.527 |
-757.487 |
DGGKM |
|
Peptide 0663 |
-7.641 |
-757.442 |
ADGLT |
|
Peptide 0252 |
-7.624 |
-757.215 |
EGGGS |
|
Peptide 1781 |
-7.577 |
-757.109 |
RDGGT |
|
Peptide 0135 |
-7.641 |
-756.293 |
AGGGG |
|
Peptide 0239 |
-7.478 |
-756.249 |
NGGSV |
|
Peptide 1527 |
-7.493 |
-755.343 |
DGGGT |
|
Peptide 0633 |
-7.5 |
-755.335 |
GGGLT |
|
Peptide 1635 |
-7.478 |
-755.334 |
GGGGS |
|
Peptide 0516 |
-7.518 |
-755.304 |
AGGGS |
|
Peptide 1069 |
-7.484 |
-755.126 |
EEGGS |
|
Peptide 0715 |
-7.471 |
-752.712 |
GGGLS |
|
Peptide 1942 |
-7.813 |
-750.946 |
NGGSY |
|
Peptide 0953 |
-7.472 |
-750.009 |
GGGLS |
|
Peptide 1707 |
-7.561 |
-749.845 |
GGGLK |
|
Peptide 1392 |
-7.549 |
-735.347 |
ARDQG |
|
Peptide 0778 |
-7.616 |
-705.833 |
NGGGT |
Figure 5: Binding conformations of top five peptide inhibitors of class D β-lactamases. Visualization of top five peptides inhibitors (A-E) in the active site of class D β-lactamases. (A) Peptide 0038 (B) Peptide 0285 (C) Peptide 1974 (D) Peptide 1183 (E) Peptide 0609. The peptides are depicted in stick representation emphasizing their spatial arrangements and binding poses
Focusing on the top five peptides from our dataset, these sequence patterns give information about the structural and functional features of peptides. All five peptides have alanine (Ala) which is known for its hydrophobic property. This feature improves the stability of peptide structure through allowing hydrophobic interaction. Also, the dipeptide glycine (Gly) appeared at many places, which allows several binding sites with more flexibility due to its small size. The polar amino acid residues (e.g. serine [Ser] and threonine [Thr]) suggest the formation of hydrogen bonds, possibly enhancing specific interactions with target proteins. These peptides are also a mixture of hydrophobic (e.g., Ala, Leu, Met) and polar residues (e.g., Ser, Thr), indicative of an intent to enhance binding in diverse environments. Moreover, cationic residues such as aspartic acid (Asp) and arginine (Arg), which or are present in many peptides, allow electrostatic interactions with negatively charged or polar targets. The existence of these common motifs indicate an intentional design of peptide sequences to enhance their binding interactions with specific proteins. With the presence of flexible glycine, hydrophobic alanine and polar or charged residues, there is greater potential for forming strong-specific bonds. The high level of glycine results in conformational changes which allows the peptides to conform to various binding sites. Alanine scanning also enriches non-covalent bonding interactions, as repeated usage of alanine strengthens hydrophobic interactions, whereas polar and charged residues promote opportunities for hydrogen bonding and electrostatic interactions to significantly increase binding affinity [24, 27, 37]. Overall, these patterns suggest that the peptides are relatively fit for use in drug design and protein-protein interaction studies since they exhibit distinct binding features necessary for effectiveness.
Moreover, the β-lactamases from different groups are characterized by extensive repeats of individual amino acids suggesting the existence of common patterns and/or motifs. The penta-peptide of interest, is enriched in peptides predicted to belong to Class C and D, such as peptide 1183, peptide 1139 and peptide 1065. This suggests that the peptides may be actually relevant for its structure or function. In addition, glycine (G) and alanine (A) have made frequent appearances in various sequences. The repeated motif AGG is consistent with a conserved function, and AAA in addition to (AGG) are found within Class B, indicating similar structural functions. The ratio between non-polar and polar residues varies across the classes. Class A and C peptides have a higher representation of hydrophobic residue (Gly(G) or Ala(A)). In contrast, in its sequence Classes B and D have a higher number of charged residues (lysine (K) and aspartate (D)). We expect that the shift in distribution of residues may alter peptide interactions and thereby stability. Prominent peptides also tend to have high content of hydrophobic residues like glycine (G) and alanine (A), which possibly provide structural stability to these peptides. In contrast, Class B could have residues like permanent positively or negatively charged amino acid residues lysine (K) and aspartic acid (D), associated with functions such as enzyme activity or substrate binding. As conserved motifs proposed to contribute significantly to the biological activity of these types of enzymes, they underscore the extent to which proper arrangements of amino acids dictate a functional role [28, 35]. Such studies might help in understanding the relevance of these patterns to β-lactamase activity and resistance mechanisms.
Conclusion
This study designed and screened for peptides targeting β-lactamases and identified candidates showing promising affinities with good inhibitory potency. Utility of these peptides in strategies to counteract antibiotic resistance thus is presented and lays the foundation for experimental validation and further development for therapy.
Funding
No external funding was received for the conduct of this study
Author contributions
All authors approves and confirms sole responsibility for this work
Conflict of interest
The authors declares no conflicts of interest
References
- Prestinaci F, Pezzotti P, Pantosti A. Antimicrobial resistance: a global multifaceted phenomenon. Pathogens and Global Health 109 (2015): 309-18.
- Tooke CL, Hinchliffe P, Bragginton EC, Colenso CK, Hirvonen VHA, et al. β-Lactamases and β-Lactamase Inhibitors in the 21st Century. Journal of Molecular Biology 431 (2019): 3472-3500.
- Murray CJL, Ikuta KS, Sharara F, Swetschinski L, Aguilar GR, et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. The Lancet 399 (2022): 629-55. 0
- Medeiros I, Aguiar AJFC, Fortunato WM, Teixeira AFG, Silva EGOE, et al. In silico structure-based design of peptides or proteins as therapeutic tools for obesity or diabetes mellitus: A protocol for systematic review and meta analysis. Medicine 102 (2023): e33514.
- Hashemi ZS, Zarei M, Fath MK, Ganji M, et al. In silico Approaches for the Design and Optimization of Interfering Peptides Against Protein-Protein Interactions. Frontiers in Molecular Biosciences 8 (2021).
- Badhe Y, Gupta R, Rai B. In silico design of peptides with binding to the receptor binding domain (RBD) of the SARS-CoV-2 and their utility in bio-sensor development for SARS-CoV-2 detection. RSC Advances 11 (2021): 3816-26.
- Fiser A, Šali A. Modeller: Generation and Refinement of Homology-Based Protein Structure Models. Methods in Enzymology on CD-ROM/Methods in Enzymology (2003): 461-91.
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-Pdb Viewer: An environment for comparative protein modeling. Electrophoresis 18 (1997): 2714-23.
- Laskowski RA, MacArthur MW, et al, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. Journal of Applied Crystallography 26 (1993): 283-91.
- Eisenberg D, Lüthy R, Bowie JU. VERIFY3D: Assessment of protein models with three-dimensional profiles. Methods in Enzymology on CD-ROM/Methods in Enzymology (1997): 396-404.
- Colovos C, Yeates TO. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Science 2 (1993): 1511-9.
- Bernhofer M, Dallago C, Karl T, Satagopam V, Heinzinger M, Littmann M, et al. PredictProtein - Predicting Protein Structure and Function for 29 Years. Nucleic Acids Research 49 (2021): W535-40.
- McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics 16 (2000): 404-5.
- Wallner B, Elofsson A. Identification of correct regions in protein models using structural, alignment, and consensus information. Protein Science 15 (2006): 900-13.
- McGuffin LJ. The ModFOLD server for the quality assessment of protein structural models. Bioinformatics 24 (2008): 586-7.
- Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Research 35 (2007): W407-10.
- Barnsley KK, Ondrechen MJ. Enzyme active sites: Identification and prediction of function using computational chemistry. Current Opinion in Structural Biology 74 (2022): 102384.
- Binkowski TA. CASTp: Computed Atlas of Surface Topography of proteins. Nucleic Acids Research 31 (2003): 3352-5.
- Zu-Kang F, Sippl MJ. Optimum superimposition of protein structures: ambiguities and implications. Folding and Design 1 (1996): 123-32.
- Kabsch W, Sander C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22 (1983): 2577-637.
- Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry 31 (2009): 455-61.
- Kaufmann KW, Lemmon GH, DeLuca SL, et al. Practically Useful: What the RosettaProtein Modeling Suite Can Do for You. Biochemistry 49 (2010): 2987-98.
- Rohl CA, Strauss CEM, Misura KMS, Baker D. Protein Structure Prediction Using Rosetta. Methods in Enzymology on CD-ROM/Methods in Enzymology (2004): 66-93.
- Gagat P, Ostrówka M, Duda-Madej A, et al. Enhancing Antimicrobial Peptide Activity through Modifications of Charge, Hydrophobicity, and Structure. International Journal of Molecular Sciences 25 (2024): 10821
- O’Meara MJ, Leaver-Fay A, Tyka MD, Stein A, et al. Combined Covalent-Electrostatic Model of Hydrogen Bonding Improves Structure Prediction with Rosetta. Journal of Chemical Theory and Computation 11 (2015): 609-22.
- Aledo JC. Methionine in proteins: The Cinderella of the proteinogenic amino acids. Protein Science 28 (2019): 1785-96
- Van Rosmalen M, Krom M, Merkx M. Tuning the Flexibility of Glycine-Serine Linkers to Allow Rational Design of Multidomain Proteins. Biochemistry 56 (2017): 6565-74.
- Datta A, Jaiswal N, Ilyas H, Debnath S, et al. Structural and Dynamic Insights into a Glycine-Mediated Short Analogue of a Designed Peptide in Lipopolysaccharide Micelles: Correlation Between Compact Structure and Anti-Endotoxin Activity. Biochemistry 56 (2017): 1348-62.
- Zhang S, Gangal G, Uludağ H. ‘Magic bullets’ for bone diseases: progress in rational design of bone-seeking medicinal agents. Chemical Society Reviews 36 (2006): 507-31.
- Browning NJ, Perez MAS, Brunk E, Rothlisberger U. EVOLVE: An Evolutionary Toolbox for the Design of Peptides and Proteins. ChemRxiv (2023).
- Unnikrishnan AC, Das BK, Saveri P, Mani E, et al. Efficiency Enhancement in Peptide Hydrogelators: The Crucial Role of Side Chain Hydrogen Bonding Over Aromatic π-π Interactions. Langmuir (2024).
- Wang B, Xie N, Li B. Influence of peptide characteristics on their stability, intestinal transport, and in vitro bioavailability: A review. Journal of Food Biochemistry 43 (2018): e12571.
- Rahban M, Zolghadri S, Salehi N, Ahmad F, Haertlé T, et al. Thermal stability enhancement: Fundamental concepts of protein engineering strategies to manipulate the flexible structure. International Journal of Biological Macromolecules 214 (2022): 642-54.
- Gupta MN, Uversky VN. Biological importance of arginine: A comprehensive review of the roles in structure, disorder, and functionality of peptides and proteins. International Journal of Biological Macromolecules 257 (2023): 128646.
- Van Rosmalen M, Krom M, Merkx M. Tuning the Flexibility of Glycine-Serine Linkers to Allow Rational Design of Multidomain Proteins. Biochemistry 56 (2017): 6565-74.
- Tapken D, Steffensen TB, Leth R, Kristensen LB, et al. The low binding affinity of D-serine at the ionotropic glutamate receptor GluD2 can be attributed to the hinge region. Scientific Reports 7 (2017).
- Chowdhury R, Boorla VS, Maranas CD. Computational biophysical characterization of the SARS-CoV-2 spike protein binding with the ACE2 receptor and implications for infectivity. Computational and Structural Biotechnology 18 (2020): 2573-82.






 Impact Factor: * 6.2
Impact Factor: * 6.2 Acceptance Rate: 76.33%
Acceptance Rate: 76.33%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 