The Effectiveness of HIV Drug Resistance Mutation Testing on Dried Blood Spot (DBS) Samples in Vietnam
Tram Hong Tran1, Thien Huu Doan1, Huong Thi Thu Phan2, Xuyen Huu Doan3, Thai Duy Nguyen1, *
1National Institute for Control of Vaccines and Biological, 1 Nghiem Xuan Yem Str., Dai Kim Ward, Hoàng Mai Dist., Hanoi, 100000
2Vietnam Administration of HIV/AIDS Control, Lane 8 Ton That Thuyet Str., My Dinh Ward, Nam Tu Liem Dist., Hanoi, 100000
3Vietnam University of Traditional Medicine, 2 Tran Phu Rd., Mo Lao Ward, Ha Dong Dist., Hanoi 100000
*Corresponding author: Nguyen Duy Thai, National Institute for Control of Vaccines and Biologicals, 1 Nghiem Xuan Yem Str., Dai Kim ward, Hoàng Mai Disst., Hanoi, Vietnam, 100000
Received: 17 September 2023; Accepted: 29 September 2023; Published: 06 October 2023
Article Information
Citation: Tram Hong Tran, Thien Huu Doan, HÆ°Æ¡ng Thi Thu Phan, Xuyen Huu Doan, Thai Duy Nguyen. The Effectiveness of HIV Drug Resistance Mutation Testing on Dried Blood Spot (DBS) Samples in Vietnam. Fortune Journal of Health Sciences. 6 (2023): 363-372.
View / Download Pdf Share at FacebookAbstract
Background: Over the past years, numerous countries worldwide have conducted investigations into HIV drug resistance (HIVDR) using methods recommended by the World Health Organization (WHO) [1]. In 2017, WHO launched a global action plan on HIV drug resistance, outlining strategies for prevention, monitoring, and response to HIV drug resistance in order to achieve the goal of controlling and ultimately ending the HIV pandemic by 2030 [2, 3].
Objective: To evaluate the effectiveness of various diagnostic kits used in genetic testing to detect HIV drug-resistant mutations on dried blood spot (DBS) samples in Vietnam. Method: 56 research articles were selected following PRISMA guidelines for the study.
Results: Globally, four types of diagnostic kits—Trugene, Viroseq, ATCC, and In-house nested PCR—are employed for genetic sequencing to identify drug-resistant mutations on DBS samples [4, 5]. With advantages such as ease of implementation, small sample volume (100 ul/drop), cost-effectiveness, convenience in sample transportation at room temperature, and reduced risk of cross-contamination, DBS has gradually replaced plasma samples in general molecular biological tests and genetic sequencing tests in particular [6].
Conclusion: The In-house nested RT-PCR technique can be used for testing HIV drug-resistant mutations on DBS samples with HIV viral loads greater than 1000 copies/ml in Vietnam, as an alternative to plasma samples [7].
Keywords
<p>HIV drug resistance mutation test, DNA sequensing, DBS sample, In-housed nested RT-PCR</p>
Article Details
1. Introduction
Worldwide, dried blood spot (DBS) samples have been extensively studied for early diagnosis in children under 18 months suspected of HIV infection [8, 9], measuring HIV viral load (9-19) and identifying drug-resistant HIV mutation genotypes [11, 20-45]. However, DBS contains both pre-viral DNA and cell RNA, which can lead to false positives, particularly in cases of plasma samples with very low viral loads or undetectable. Therefore, the World Health Organization (WHO) recommends cautious consideration when using DBS for viral load testing [11, 19]. In addition, the WHO has collaborated with reputable testing laboratories to develop, confirm, and standardize methods for detecting drug-resistant HIV mutations from DBS samples [15].
In Vietnam, the method of collecting samples on dried blood spot papers is widely applied in medical practice for diagnosing infectious diseases, metabolic disorders, hereditary conditions, and more in newborns and young children due to its advantages in terms of time, convenience in sampling, preservation, and sample transportation. It has also been used for early diagnosis of HIV infection in children under 18 months since 2009 [46], measuring HIV viral load from DBS samples since 2016 [47-50] and conducting genetic sequencing studies on DBS samples compared to plasma samples to identify drug-resistant HIV mutations since 2012 [51].
Testing for HIV-1 drug resistance mutations is an essential part of WHO's global drug resistance evaluation and prevention strategy [52, 53]. Plasma samples are always considered the gold standard for identifying the HIV drug resistance genotype. However, the use of plasma samples faces many challenges in terms of personnel and equipment needed to preserve the samples, especially in impoverished rural areas, remote and isolated regions, particularly in low- and middle-income countries. Therefore, dried blood spot (DBS) samples have been and are being used as a replacement for plasma samples in determining the HIV drug resistance genotype. The DBS sampling technique is simple, with a small sample volume (100ul/drop), can be stored at room temperature, has minimal cross-contamination, is easy to transport, requires no special laboratory handling, and can be used for routine clinical or surveillance purposes [54]. Mac dù có nhung ?u diem nhu vay nhung DBS van có mot so nh??c diem, truoc het là do nhay cua Despite these advantages, DBS has some limitations, primarily the sensitivity of the viral RNA amplification process is less than with plasma samples due to the smaller volume. Additionally, for patients with a low viral load, the genotype results from DBS samples may not accurately reflect the current drug resistance mutations, so it is recommended to use plasma samples for testing in these cases. The virus's RNA in DBS can degrade if the DBS gets moist or is stored at room temperature for an extended period (> 14 days), it should be kept in a deep freezer (-70°C) immediately after the sample dried and before testing with this case. Thus, our research aims to compile and evaluate the efficacy of various HIV drug resistance mutation testing products on DBS samples compared to traditional plasma samples. This would allow for the selection of appropriate testing kits for detecting HIV drug resistance mutations in DBS samples in mountainous areas, remote regions, borders, islands, and other areas in Vietnam where the conditions for using other plasma samples are not met.
2. Method
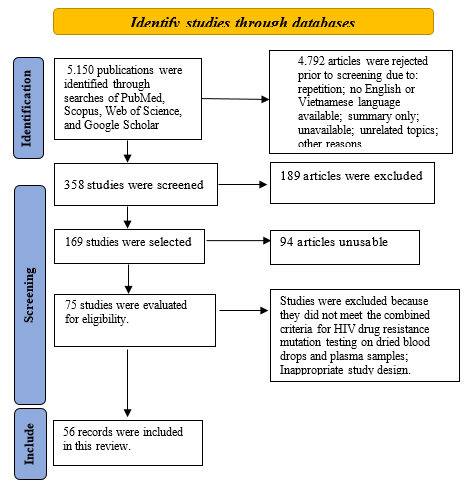
We conducted a search for open-access articles on databases including PubMed, Scopus, Web of Science, and Google Scholar from 2006 to the present using the keywords: Drug resistance mutation testing; HIV gene sequencing assays; HIV testing by dried blood spot (DBS); HIV testing on plasma samples. A total of 5,150 articles were found for these keywords. After filtering based on criteria that combined HIV drug resistance mutation testing on dried blood spot samples and plasma samples, we selected 56 relevant reports for analysis in this study. The protocol for this assessment follows the PRISMA guidelines.
3. Results
3.1. Evaluation of HIV drug resistance mutation testing products on DBS samples
Since 2004, following the recommendation of WHO, low and middle-income countries have optimized techniques and implemented dried blood spot (DBS) samples in HIV drug resistance mutation testing. By 2020, four main products were used worldwide (Table 2) [10, 12-14, 20-25, 27-29, 35-39].
Table 1: Global studies on HIV gene testing techniques using dried blood spot (DBS)
|
No. |
Publications |
Lab Techniques |
Size of amplified gene |
Storage conditions |
Sample characteristics |
|
1 |
Bertagnolio et al. (20) |
In-house nested RT-PCR |
RT: 700 bp |
37°C; 85% humidity; 3 months |
Subjects untreated in Mexico, subtype B |
|
2 |
Buckton et al. (21) |
In-house nested RT-PCR |
protease: 758 bp RT: 805 bp |
-20°C |
Clinic patient from UK, subtypes A, C, CRF02 |
|
3 |
Garrido et al. (22) |
In-house nested RT-PCR: RT và gp41fragments |
RT: 726 bp |
4°C; no desiccant bag |
Treated patient from Angola, multiple subtypes |
|
4 |
Hallack et al. (23) |
TruGene |
1.3 kb |
-20°C |
Treated and untreated patients in the US, subtype B |
|
5 |
Masciotra et al. (24) |
ViroSeq™ |
1.8 kb |
-20°C; 18-26 weeks |
Most treated, subtype B |
|
6 |
McNulty et al. (25) |
In-house nested RT-PCR |
1 kb |
-20°C; 2-3 years |
Untreated, subtype from Cameroon, subtype A, CRF02 |
|
7 |
Steegen et al. (27) |
In-house nested RT-PCR |
protease: 458 bp RT: 646 bp |
-20°C |
Treated and untreated patients in Kenya, subtypes A, C, D, CRF16 |
|
8 |
Youngpairoj et al. (28) |
ViroSeq™ or in-house nested RT-PCR |
1.8 kb hoac 1 kb |
4°C; 1 year |
Treatment-experienced patients, subtype B |
|
9 |
Ziemniak et al. (29) |
In-house nested RT-PCR |
RT: 663 bp |
Normal temperature; 5 months |
Treated and untreated patients in the US, subtype B |
|
10 |
Zhou et al. (35) |
In-house nested RT-PCR |
1084 bp |
Normal temperature; 5 months (Nigeria) or -70°C (Vietnam) |
Treated and untreated patients in Nigeria and Vietnam |
|
11 |
Yang et al. (36) |
In-house nested RT-PCR |
1062 bp |
-20°C or -70°C |
Treated and untreated patients in China, Malawi and the Republic of Tanzania |
|
12 |
Monleau et al. (10) |
In-house nested RT-PCR (ANRS) |
protease: 507 bp RT: 798 bp |
Room temperature; 2-4 weeks; -20°C, 16-38 days |
Treated patients are from Burkina Faso, Cameroon, Senegal, Thailand, Togo and Vietnam |
|
13 |
Inzaule et al. (12) |
In-house nested RT-PCR (CDC Hoa Ky) |
1084 bp |
-30°C |
HIV-1 infected mothers and children in Kenya (Kisumu) |
|
14 |
Bronze et al. (13) |
In-house nested RT-PCR |
RT: 591 bp |
-20°C; 13-21 months |
People with HIV-1 in South Africa |
|
15 |
Aitken et al. (14) |
In-house nested RT-PCR |
RT: 591 bp |
No information |
People with HIV-1 in South Africa |
|
16 |
Monleau et al. (37) |
In-house nested RT-PCR |
RT: 798 bp |
Normal temperature (+20°C) or (+37°C); high humidity |
The patient was infected with HIV-1 in France |
|
17 |
Salimo et al. (38) |
In-house nested RT-PCR |
1084 bp |
-80°C |
HIV-infected children younger than 2 years old |
|
18 |
Zhang et al. (39) |
ATCC kitb |
1084 bp |
Normal temperature; < 2 weeks then put in -80oC |
Adults and children treated in Kenya and the Republic of Tanzania |
The results in Table 1 indicate that there are four main products used globally for HIV drug resistance mutation testing: TruGene, ViroSeq™, ATCC kit, and In-house nested RT-PCR. These products are designed to sequence the Pol gene region, encoding the amino acid substitutions known to cause ARV drug resistance[7]. The products TruGene, ViroSeq™, and ATCC kit (Thermo Fisher) are commercial offerings, of which TruGene is no longer available in the market due to its outdated equipment. Given that countries with high HIV rates are predominantly poor, the cost of these products is a significant concern. Therefore, to align with economic conditions, the In-house nested RT-PCR technique has been researched and widely applied for gene sequencing to detect HIV drug resistance mutations [10, 12-14, 20-22, 25-28, 35-38]. Out of the 18 studies listed in the table, 15 utilized the In-house nested RT-PCR technique. In Vietnam, DBS samples are stored at room temperature for 2 to 4 weeks and then preserved at -20°C until testing. Treatment failure samples are selected for sequencing to detect HIV drug resistance using the In-house nested RT-PCR method provided by the French National Agency for Research on AIDS and Viral Hepatitis (ANRS) [10].
3.2 Effectiveness assessment of DBS samples compared to plasma samples
For DBS samples, in order to have a foundation for widespread and convenient deployment in the field, experts have conducted several tests to study the stability of DBS samples when stored at different temperatures. According to the WHO summary in 2020, 8 studies have been carried out (Table 3) [7].
Table 2: Stability of DBS samples at various temperatures
|
No. |
Publications |
Storage conditions |
Moisture-proof package |
Results |
|
1 |
Bertagnolio et al. [20] |
3 months at +37°C and 85% humidity |
Yes |
Good amplification rate (90%) |
|
2 |
McNulty et al. [25] |
6 years at -30°C; 5 years at normal temperature and -70°C; 2-3 years at -20°C |
Yes |
Samples are completely damaged at room temperature; stable at -30°C and -70°C; Recommended storage for 2-3 years at -20°C. |
|
3 |
Nelson et al. [31] |
3-6 years at normal temperature |
Yes |
Average amplification success rate (69%); 1 log difference for viral load. |
|
4 |
Garcia-Lema et al. [32] |
1-16 weeks at +37oC high humidity; -20oC |
Yes |
DBS is stable at +37oC for 1-2 weeks and -20oC for a long time |
|
5 |
Monleau et al. [37] |
1-2 months at normal temperature and +37°C; high humidity |
Yes |
11/12 amplified DBS samples were stored at room temperature for 1 or 2 months; 10/12 samples or 7/12 samples at +37°C. |
|
6 |
Parry et al. [42] |
2-4 weeks at normal temperature |
Yes |
There was no sign of a decrease in amplification rates for samples at room temperature: for 2 weeks (93%) compared to frozen or plasma samples (97-98%); decreased slightly at 4 weeks (89%). |
|
7 |
Wallis et al. [43] |
3 months at normal temperature, +4°C and -20°C |
Yes |
Some samples decreased in amplification when stored at room temperature compared to the same sample stored at +4°C and -20°C |
|
8 |
Aitken et al. [44] |
4 weeks at -20°C and +30°C |
Yes |
Stable at -20°C and +30°C (viral load > 10.000 copies/ml) for 2 weeks; Slightly reduced at 2 weeks, +30°C (viral load: 1.000 copies/ml). |
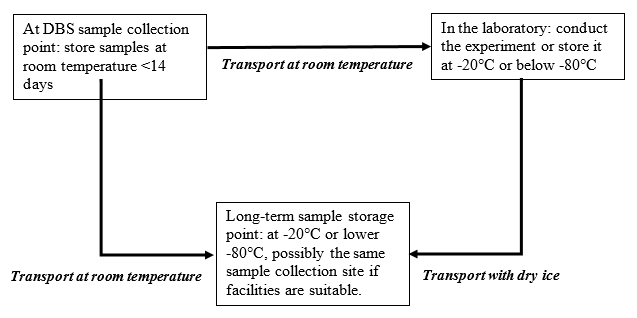
Research results from Table 2 indicate that the sample remains stable when stored at room temperature, or at 4°C for up to 2 weeks, or at -20°C for 2-3 years, or at -30°C and -70°C for extended periods. Based on these studies, the WHO has provided guidelines for transporting DBS samples [7]. In Vietnam, following the success of the research on DBS samples [47-49], the Ministry of Health issued Decision 1112/QD-BYT providing guidelines for conducting HIV load testing in monitoring and treating HIV/AIDS. Specifically, in Annex 3B, the procedure for sampling, storing, packing, and transporting DBS samples for HIV load testing was stipulated, similar to the WHO's procedure as below [55].
|
At DBS sample collection point: store samples at room temperature <14 days |
|
In the laboratory: conduct the experiment or store it at -20°C or below -80°C |
|
Long-term sample storage point: at -20°C or lower |
|
Transport at room temperature |
|
Transport at room temperature |
|
Transport with dry ice |
Research indicates that DBS sampling is simple, manpower-efficient, labor-saving, and convenient for transportation. It can be transported at normal temperatures without a complicated cold chain like plasma sample storage. Moreover, it also doesn't break and cause cross-contamination. From a review of 18 studies worldwide on gene sequencing tests to detect HIV drug resistance mutations, eight studies did not compare mutation locations between DBS samples and plasma samples but were conducted directly on HIV-positive DBS samples [13, 21, 22, 27, 29, 36, 37, 39]. The remaining ten studies compared mutation locations between DBS samples and plasma samples, as shown in Table 4 [10, 12, 14, 20, 23-25, 28, 35, 36, 38].
Results in Table 3 indicate that, depending on the specific product and chemicals used, the sensitivity in detecting mutations varies at different HIV viral load thresholds. All studies reported results that for samples with a viral load > 1.000 copies/ml, the amplification success rate is higher than for those with a viral load < 1.000 copies/ml (>70%). Consequently, the WHO has recommended that for DBS samples, the minimum viral load threshold should be 1000 copies/ml (with the viral load value taken from measurements on plasma samples) for sequencing tests to detect HIV drug resistance mutations [7]. In Vietnam, the threshold for treatment failure in HIV is set at ≥ 1.000 copies/ml, as stipulated in professional guidelines and legal documents [55].
Table 3: Amplification success rate with HIV-positive DBS sample
|
No. |
Studies |
Number of samples |
Viral load (copy/ml) |
Amplification success rate on DBS sample |
|
1 |
Buckton et al. [21] |
12 |
80 to 115.300 |
Protease: 83% RT: 100% |
|
2 |
Garrido et al. [22] |
77 |
1.000 to 850.000 |
RT: 30%; gp41: 43% |
|
3 |
Steegen et al. [27] |
29 |
55 to >100.000 |
96% either protein region or RT region, 89.7% both gene regions, viral load (>100 copies/ml): 100% |
|
4 |
Ziemniak et al. [29] |
9 |
<50 to 94.600 (average: 17.792) |
Total: 94%, viral load (193 copies/ml): 100% |
|
5 |
Yang et al. [36] |
171 |
70 samples: <400 to 367.875 (average: 8.021) |
Total: 87.1%; 70 samples: 94% |
|
6 |
Bronze et al. [13] |
41 |
270 to 15.519.576 |
76% for viral loads >5.000 copies/ml; 15% for viral loads <5000 copies/ml. |
|
7 |
Monleau et al. [37] |
12 |
1.380 to 263.000 |
100% |
|
8 |
Zhang et al. [39] |
499 |
>1.000 |
92%; ~82% for viral loads 1.000-5.000 copies/ml |
Table 4: Comparison of gene sequencing between DBS samples and plasma samples
|
No. |
Studies |
Number of samples |
Viral load (copy/ml) |
Amplification success rate on DBS sample |
% sequence homology compared to plasma |
|
1 |
Bertagnolio et al. [20] |
103 |
No testing |
90.1% either protein region or RT region; 78.2% both gene regions. |
99% |
|
2 |
Hallack et al. [23] |
33 |
1.178 to 414.212 (average: 11.666) |
Total: 78.8%; viral load >60.000 copies/ml: 90.5%; viral load <60.000 copies/ml: 58.3% |
99.30% |
|
3 |
Masciotra et al. [24] |
60 |
78 to 676.694 (average: 9.135) |
Total: 83%, viral load >2000 copies/ml: 100%; viral load <2000 copies/ml: 54% |
98.80% |
|
4 |
McNulty et al. [25] |
40 |
665 to 645.256 (average: 23.715 |
Total: 92%; viral load >10.000 copies/ml: 100%; viral load <10.000 copies/ml: 73% |
98.50% |
|
5 |
Youngpairoj et al. [28] |
40 |
518 to 676.694 (average: 13.680) |
ViroSeq™: 57.5%; In-house: 95% |
94.50% |
|
6 |
Zhou et al. [35] |
98 |
Nigeria: 150 to 436.500 (average: 9.332) |
Nigeria: 96.1%; |
Nigeria: 98.8%; |
|
Vietnam: 95.8% |
Vietnam: 98.9% |
||||
|
7 |
Monleau et al. [10] |
124 |
≥ 1.000 plasma samples |
77% |
78% |
|
8 |
Inzaule et al. [12] |
68 |
>10.000: n = 44; 1000-10.000: n = 13 |
>10.000 copies/ml: 100%; |
99.50% |
|
1.000 - 10.000 copies/ml: 54% |
|||||
|
9 |
Aitken et al. [14] |
25 |
1020 to 449.000 |
84% |
97% |
|
10 |
Salimo et al. [38] |
238 |
Mostly >100.000 |
98.70% |
99.50% |
Table 4 shows that in 10 studies comparing the similarity in sequencing success rates between DBS samples and plasma samples, the results are equivalent, averaging about 97%. This provides a basis for underdeveloped countries to widely implement HIV molecular biological testing. In Vietnam, there are two studies comparing the genetic sequencing results between DBS and plasma. One by Monleau et al reported a 78% similarity in mutation positions [10]. Meanwhile, the study by Zhou et al showed a similarity rate of up to 98.9% in mutation positions [35].
4. Discussion
Results in Table 2 showed three utilized commercial products TruGene, ViroSeq™, and the ATCC kit (Thermo Fisher). The remaining 15 studies used the In-house nested RT-PCR technique. These studies were primarily conducted in poorer, underdeveloped countries such as Africa, Vietnam, Thailand, etc. With a high number of HIV-infected individuals and low economic status, especially in African regions, there is a scarcity of medical manpower and equipment. Thus, using commercial products to detect HIV drug resistance is costly and challenging. Hence, implementing drug resistance mutation testing using the In-house nested RT-PCR technique is more fitting for these underdeveloped countries' realities [10, 12-14, 23, 27, 35, 36, 39].
Plasma samples have always been considered the gold standard in HIV molecular biological tests. However, since 2009, the world has researched the use of dried blood spots (DBS) for early diagnostic testing of HIV in children under 18 months using PCR technology. WHO recommended its use in countries with high HIV prevalence such as Africa, Vietnam, Thailand, etc [8, 46]. Manufacturers like Abbott, Roche, and others have since developed commercial kits to expand its use for HIV viral load testing and resistance mutation detection in DBS samples. Yet, implementing DBS for viral load testing and seeking resistance mutations in HIV has faced challenges, especially for samples with low viral loads of <1000 copies/ml [35]. In the study by Zhou et al., there was a focus on optimizing genotypic testing using the low-cost In-house technique to monitor and track HIV drug resistance in underdeveloped countries with high HIV prevalence such as Africa, Vietnam, Thailand, etc [35]. Additionally, the research also explored the application of DBS samples in drug resistance testing as an alternative to plasma samples. The study of Zhou et al. had indicated [35].
The optimized In-house testing method demonstrated high sensitivity suitable for various HIV-1 subtypes such as M and CRF groups were carried out on both plasma and DBS samples collected from six countries with high HIV prevalence, namely Cameroon, Malawi, Nigeria, Zambia, Thailand, and Vietnam. This In-house testing, when compared with commercial kits like ViroSeq™ and Trugene, yielded equivalent results, with respective accuracies of 99.3%, 99.6%, and 99.1%. Moreover, the research team estimated that the reagent cost per test for the In-house method is 40 USD, compared to 213.20 USD for Trugene and 172.86 USD for ViroSeq™. Hence, both in terms of technical sensitivity and economy, the In-house nested PCR technique can be widely implemented for HIV drug resistance testing in underdeveloped countries for patients suspected of treatment failure with viral loads >1000 copies/ml.
On the other hand, the study successfully compared the results of genotypic sequencing to identify drug resistance mutations between DBS samples and plasma samples. The HIV genotypic sequencing results on 18 out of 26 DBS samples compared to their corresponding plasma samples collected from virologically failing patients in Nigeria and 69 out of 72 DBS samples compared to their corresponding plasma samples collected from patients suspected of treatment failure in Vietnam were identical in terms of drug resistance mutation points. Notably, the 18 DBS samples from Nigeria were stored at room temperature for an average of 85 days before being transferred to the laboratory. Additionally, two external quality control DBS PT samples, whether shipped frozen or at ambient temperature, resulted in successful amplification and genotypic sequencing, except for one sample with the lowest HIV load (<1.000 copies/ml) that was transported at ambient temperature. These results suggest that the optimized In-house nested PCR test has high sensitivity in determining the genotype for both plasma and DBS samples. Hence, the study indicates that DBS samples remain stable even when stored at room temperature for extended periods. However, they need to be meticulously packaged and stored in an air-conditioned room with low humidity (possibly with the aid of a dehumidifier) to ensure better sample quality. Proper packaging and storage of DBS are crucial factors to guarantee successful genotyping results. The research also suggests that for samples with low viral loads, the packaging and transportation must be even stricter to maintain sample quality. In cases where the DBS sample cannot be amplified, it is imperative to use plasma samples.
Another study by Zhang et al. indicated that DBS samples have advantages such as not requiring expensive equipment, easy for training, not necessitating cold chain storage, and not contaminating the surrounding environment, etc (39). The study also estimated that the cost of transporting DBS samples is about 250 USD depending on the country, while the estimated cost of using plasma samples is approximately 5,000 USD. Furthermore, the use of the ATCC kit (essentially a commercialized version of the In-house nested RT-PCR by Thermo Fisher) also reduces the cost of determining the HIV drug resistance genotype. Other HIV drug resistance genotyping products available in the market cost around 200 USD per test at the time of the study, while the cost of testing with the ATCC kit is about 60 USD per test. For 500 samples genotyped for HIV, just the cost of the drug resistance testing kit alone saves 50.000 USD. Based on these credible research results, WHO has recommended that developing countries like Vietnam use the In-house nested RT-PCR technique on DBS samples for HIV drug resistance mutation testing, as per the WHO 2020 guidelines [7].
5. Conclusion
The In-house test ensures efficiency and cost savings in identifying drug-resistant HIV genotypes compared to commercial kits. DBS samples are cost-effective, easy to train, and do not require cold storage like plasma samples. Also, according to WHO recommendations, dried DBS samples are safe and non-infectious, making them suitable for areas with limited medical resources. Thus, using the In-house technique on DBS samples is effective for HIV testing to mitigate the growth and transmission of drug-resistant HIV in the community. However, for samples with a viral load of less than 1,000 copies/ml, plasma samples are the best choice.
References
- World Health Organization. HIV drug resistance report 2019.
- World Health Organization. Global Action Plan on HIV drug resistance 2017–2021.
- Bertagnolio S, Beanland RL, Jordan MR, Doherty M, Hirnschall G. The World Health Organization’s response to emerging human immunodeficiency virus drug resistance and a call for global action. The Journal of infectious diseases 216 (2017): S801-S4.
- World Health Organization. HIV drug resistance report 2021.
- World Health Organization. HIV drug resistance strategy 2021.
- Malsagova K, Kopylov A, Stepanov A, Butkova T, Izotov A, Kaysheva A. Dried blood spot in laboratory: directions and prospects. Diagnostics 10 (2020): 248.
- World Health Organization. WHO manual for HIV drug resistance testing using dried blood spot specimens.
- Stevens W, Sherman G, Downing R, Parsons L, Ou C-Y, Crowley S, et al. Role of the laboratory in ensuring global access to ARV treatment for HIV-infected children: consensus statement on the performance of laboratory assays for early infant diagnosis. The open AIDS journal 2 (2008): 17.
- Smit PW, Sollis KA, Fiscus S, Ford N, Vitoria M, Essajee S, et al. Systematic review of the use of dried blood spots for monitoring HIV viral load and for early infant diagnosis. PLoS One 9 (2014): e86461.
- Monleau M, Aghokeng AF, Eymard-Duvernay S, Dagnra A, Kania D, Ngo-Giang-Huong N, et al. Field evaluation of dried blood spots for routine HIV-1 viral load and drug resistance monitoring in patients receiving antiretroviral therapy in Africa and Asia. Journal of clinical microbiology 52 (2014): 578-86.
- World Health Organization. HIV molecular diagnostics toolkit to improve access to viral load testing and infant diagnosis.
- Inzaule S, Yang C, Kasembeli A, Nafisa L, Okonji J, Oyaro B, et al. Field evaluation of a broadly sensitive HIV-1 in-house genotyping assay for use with both plasma and dried blood spot specimens in a resource-limited country. Journal of clinical microbiology 51 (2013): 529-39.
- Bronze M, Aitken SC, Wallis CL, Steegen K, Stuyver L, de Wit TR, et al. Evaluation of an affordable HIV-1 virological failure assay and antiretroviral drug resistance genotyping protocol. Journal of virological methods 194 (2013): 300-7.
- Aitken SC, Bronze M, Wallis CL, Stuyver L, Steegen K, Balinda S, et al. A pragmatic approach to HIV-1 drug resistance determination in resource-limited settings by use of a novel genotyping assay targeting the reverse transcriptase-encoding region only. Journal of clinical microbiology 51 (2013): 1757-61.
- Parkin N, de Mendoza C, Schuurman R, Jennings C, Bremer J, Jordan MR, et al. Evaluation of in-house genotyping assay performance using dried blood spot specimens in the Global World Health Organization laboratory network. Clinical infectious diseases 54 (2012): S273-S9.
- Hamers RL, Smit PW, Stevens W, Schuurman R, Rinke de Wit TF. Dried fluid spots for HIV type-1 viral load and resistance genotyping: a systematic review. Antiviral therapy 14 (2009): 619-29.
- Johannessen A, Garrido C, Zahonero N, Sandvik L, Naman E, Kivuyo SL, et al. Dried blood spots perform well in viral load monitoring of patients who receive antiretroviral treatment in rural Tanzania. Clinical Infectious Diseases 49 (2009): 976-81.
- Guichet E, Serrano L, Laurent C, Eymard-Duvernay S, Kuaban C, Vidal L, et al. Comparison of different nucleic acid preparation methods to improve specific HIV-1 RNA isolation for viral load testing on dried blood spots. Journal of virological methods 251 (2018): 75-9.
- Parkin NT. Measurement of HIV-1 viral load for drug resistance surveillance using dried blood spots: literature review and modeling of contribution of DNA and RNA. AIDS reviews 16 (2014): 160-71.
- Bertagnolio S, Soto-Ramirez L, Pilon R, Rodriguez R, Viveros M, Fuentes L, et al. HIV-1 drug resistance surveillance using dried whole blood spots. Antiviral therapy 12 (2007): 107-14.
- Buckton AJ, Bissett SL, Myers RE, Beddows S, Edwards S, Cane PA, et al. Development and optimization of an internally controlled dried blood spot assay for surveillance of human immunodeficiency virus type-1 drug resistance. Journal of antimicrobial chemotherapy 62 (2008): 1191-8.
- Garrido C, Zahonero N, Fernandes D, Serrano D, Silva AR, Ferraria N, et al. Subtype variability, virological response and drug resistance assessed on dried blood spots collected from HIV patients on antiretroviral therapy in Angola. Journal of antimicrobial chemotherapy 61 (2008): 694-8.
- Hallack R, Doherty LE, Wethers JA, Parker MM. Evaluation of dried blood spot specimens for HIV-1 drug-resistance testing using the Trugene® HIV-1 genotyping assay. Journal of Clinical Virology 41 (2008): 283-7.
- Masciotra S, Garrido C, Youngpairoj AS, McNulty A, Zahonero N, Corral A, et al. High concordance between HIV-1 drug resistance genotypes generated from plasma and dried blood spots in antiretroviral-experienced patients. Aids 21 (2007): 2503-11.
- McNulty A, Jennings C, Bennett D, Fitzgibbon J, Bremer JW, Ussery M, et al. Evaluation of dried blood spots for human immunodeficiency virus type 1 drug resistance testing. Journal of Clinical Microbiology 45 (2007): 517-21.
- Plantier J-C, Dachraoui R, Lemée V, Gueudin M, Borsa-Lebas F, Caron F, et al. HIV-1 resistance genotyping on dried serum spots. Aids 19 (2005): 391-7.
- Steegen K, Luchters S, Demecheleer E, Dauwe K, Mandaliya K, Jaoko W, et al. Feasibility of detecting human immunodeficiency virus type 1 drug resistance in DNA extracted from whole blood or dried blood spots. Journal of Clinical Microbiology 45 (2007): 3342-51.
- Youngpairoj AS, Masciotra S, Garrido C, Zahonero N, de Mendoza C, García-Lerma JG. HIV-1 drug resistance genotyping from dried blood spots stored for 1 year at 4 C. Journal of Antimicrobial Chemotherapy. 2008;61(6):1217-20.
- Ziemniak C, George-Agwu A, Moss WJ, Ray SC, Persaud D. A sensitive genotyping assay for detection of drug resistance mutations in reverse transcriptase of HIV-1 subtypes B and C in samples stored as dried blood spots or frozen RNA extracts. Journal of virological methods 136 (2006): 238-47.
- Monleau M, Montavon C, Laurent C, Segondy M, Montes B, Delaporte E, et al. Evaluation of different RNA extraction methods and storage conditions of dried plasma or blood spots for human immunodeficiency virus type 1 RNA quantification and PCR amplification for drug resistance testing. Journal of clinical microbiology 47 (2009): 1107-18.
- Nelson JA, Loftis AM, Kamwendo D, Fawzi WW, Taha TE, Goldenberg RL, et al. Nevirapine resistance in human immunodeficiency virus type 1-positive infants determined using dried blood spots stored for up to six years at room temperature. Journal of clinical microbiology 47 (2009): 1209-11.
- García-Lerma JG, McNulty A, Jennings C, Huang D, Heneine W, Bremer JW. Rapid decline in the efficiency of HIV drug resistance genotyping from dried blood spots (DBS) and dried plasma spots (DPS) stored at 37 C and high humidity. Journal of Antimicrobial Chemotherapy 64 (2009): 33-6.
- Dachraoui R, Depatureaux A, Chakroun M, Fodha I, Letaief A, Trabelsi A, et al. Monitoring of HIV-1 resistance in Tunisia (North Africa) with a dried plasma spot strategy. JAIDS Journal of Acquired Immune Deficiency Syndromes 47 (2008): 522-5.
- Ziemniak C, Mengistu Y, Ruff A, Chen Y-H, Khaki L, Bedri A, et al. Use of dried-blood-spot samples and in-house assays to identify antiretroviral drug resistance in HIV-infected children in resource-constrained settings. Journal of clinical microbiology 49 (2011): 4077-82.
- Zhou Z, Wagar N, DeVos JR, Rottinghaus E, Diallo K, Nguyen DB, et al. Optimization of a low cost and broadly sensitive genotyping assay for HIV-1 drug resistance surveillance and monitoring in resource-limited settings. PLoS One 6 (2011): e28184.
- Yang C, McNulty A, Diallo K, Zhang J, Titanji B, Kassim S, et al. Development and application of a broadly sensitive dried-blood-spot-based genotyping assay for global surveillance of HIV-1 drug resistance. Journal of clinical microbiology 48 (2010): 3158-64.
- Monleau M, Butel C, Delaporte E, Boillot F, Peeters M. Effect of storage conditions of dried plasma and blood spots on HIV-1 RNA quantification and PCR amplification for drug resistance genotyping. Journal of Antimicrobial Chemotherapy 65 (2010): 1562-6.
- Salimo AT, Ledwaba J, Coovadia A, Abrams EJ, Technau K-G, Kuhn L, et al. The use of dried blood spot specimens for HIV-1 drug resistance genotyping in young children initiating antiretroviral therapy. Journal of virological methods 223 (2015): 30-2.
- Zhang G, DeVos J, Medina-Moreno S, Wagar N, Diallo K, Beard RS, et al. Utilization of dried blood spot specimens can expedite nationwide surveillance of HIV drug resistance in resource-limited settings. PLoS One 13 (2018): e0203296.
- Rottinghaus E, Bile E, Modukanele M, Maruping M, Mine M, Nkengasong J, et al. Comparison of Ahlstrom grade 226, Munktell TFN, and Whatman 903 filter papers for dried blood spot specimen collection and subsequent HIV-1 load and drug resistance genotyping analysis. Journal of Clinical Microbiology 51 (2013): 55-60.
- Rottinghaus EK, Beard RS, Bile E, Modukanele M, Maruping M, Mine M, et al. Evaluation of dried blood spots collected on filter papers from three manufacturers stored at ambient temperature for application in HIV-1 drug resistance monitoring. PLoS One 9 (2014): e109060.
- Parry C, Parkin N, Diallo K, Mwebaza S, Batamwita R, DeVos J, et al. Field study of dried blood spot specimens for HIV-1 drug resistance genotyping. Journal of clinical microbiology 52 (2014): 2868-75.
- Wallis C, Bell C, Horsfield P, de Wit TR, Stevens W, editors. Affordable resistance test for Africa (ARTA): DBS storage and extraction conditions for HIV subtype C. The 5th International AIDS Society Conference on HIV Pathogenesis, Treatment and Prevention (2009).
- Aitken SC, Wallis CL, Stevens W, de Wit TR, Schuurman R. Stability of HIV-1 nucleic acids in dried blood spot samples for HIV-1 drug resistance genotyping. PLoS One 10 (2015): e0131541.
- World Health Organization. WHO HIVResNet HIV drug resistance laboratory operational framework.
- Nguyen TB, Tran HT, Nguyen VN, Phan TTH, AT. N. HIV infection in HIV-exposed children under 18 months and related factors in north and north central Vietnam in 2011–2013. Vietnam Journal of Preventive Medicine (2015).
- Nguyen TA, Tran TH, Nguyen BT, Pham TTP, Hong Le NT, Ta DV, et al. Feasibility of dried blood spots for HIV viral load monitoring in decentralized area in North Vietnam in a test-and-treat era, the MOVIDA project. PLoS One 15 (2020): e0230968.
- Tran TH, Nguyen BT, Nguyen TA, Pham TTP, Nguyen TTT, Mai HTB, et al. Dried blood spots perform well to identify patients with active HCV infection in Vietnam. Journal of Viral Hepatitis 27 (2020): 514-9.
- Taieb F, Tran Hong T, Ho HT, Nguyen Thanh B, Pham Phuong T, Viet Ta D, et al. First field evaluation of the optimized CE marked Abbott protocol for HIV RNA testing on dried blood spot in a routine clinical setting in Vietnam. PLoS One 13 (2018): e0191920.
- Taieb F, Tram TH, Ho HT, Pham VA, Nguyen HL, Pham BH, et al., editors. Evaluation of two techniques for viral load monitoring using dried blood spot in routine practice in Vietnam (French National Agency for AIDS and Hepatitis Research 12338). Open forum infectious diseases (2016).
- Duc NB, Hien BT, Wagar N, Tram TH, Giang LT, Yang C, et al. Surveillance of transmitted HIV drug resistance using matched plasma and dried blood spot specimens from voluntary counseling and testing sites in Ho Chi Minh City, Vietnam, 2007–2008. Clinical infectious diseases 54 (2012): S343-S7.
- World Health Organization. Surveillance of HIV drug resistance in children newly diagnosed with HIV by early infant diagnosis.
- World Health Organization. Guidance for sampling ART clinics in countries combining surveillance of pre-treatment HIV drug resistance and acquired HIV drug resistance at 12 and 48+ months. World Health Organization (2017).
- IATA dangerous goods regulations Montreal.
- Ministry of Health Vietnam. Decision No. 1112/QD-BYT dated 26 March 2019 of the Ministry of Health on promulgating guidelines for HIV load testing in HIV/AIDS monitoring and treatment (2019).
- World Health Organization. Guidance on regulations for the Transport of Infectious Substances 2013– 2014.


 Impact Factor: * 6.2
Impact Factor: * 6.2 Acceptance Rate: 76.33%
Acceptance Rate: 76.33%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 