Early Relapse Detection and Monitoring Disease Status in Patients with Early-stage Pancreatic Adenocarcinoma using Circulating Tumor DNA
Maen Abdelrahim1,2,3*, Abdullah Esmail1, Jiaqiong Xu4, Tiffany A Katz5, Shruti Sharma5, Ekaterina Kalashnikova5, Meenakshi Malhotra5, Perry Olshan5, Paul R Billings5, Alexey Aleshin5
1Section of GI Oncology, Department of Medical Oncology, Houston Methodist Cancer Center, Houston, TX, United States
2Cockrell Center of Advanced Therapeutics Phase I program, Houston Methodist Research Institute, Houston, TX, United States
3Weill Cornell Medical College, New York, NY, United States
4Center for Outcomes Research, Houston Methodist Research Institute, Houston, TX United States.
5Natera, Inc., San Carlos, CA, United States
*Corresponding author: Maen Abdelrahim, MD, PhD, Pharm.B. Associate Professor, Section of GI oncology, Department of Medical Oncology, Houston Methodist Cancer Center. Weill Cornell Medical College and Cockrell Center of Advanced Therapeutics Phase I program. 6445 Fannin, OPC-24, Houston, TX 77030, United States
Received: 17 October 2021; Accepted: 26 October 2021; Published: 02 November 2021
Article Information
Citation: Maen Abdelrahim, Abdullah Esmail, Jiaqiong Xu, Tiffany A Katz, Shruti Sharma, Ekaterina Kalashnikova, Meenakshi Malhotra, Perry Olshan, Paul R Billings, Alexey Aleshin. Early Relapse Detection and Monitoring Disease Status in Patients with Early-stage Pancreatic Adenocarcinoma using Circulating Tumor DNA. Journal of Surgery and Research 4 (2021): 602-615.
View / Download Pdf Share at FacebookAbstract
Pancreatic ductal adenocarcinoma (PDAC) has one of the most aggressive cancer histologies, with high recurrence (85%) and a 5-year survival rate of 9%. The circulating tumor DNA (ctDNA) is evolving field in setting of management for patients with advanced solid tumors. In this study, our aim was to investigate the potential contributions of ctDNA in early-stage pancreatic adenocarcinoma prognostication as early relapse detection and disease status monitoring and to evaluate the relationship with tumor markers carcinoembryonic antigen (CEA) and cancer antigen 19-9 (CA 19-9).
Methods: We enrolled 9 patients with pancreatic cancer from whom we collected paired tissue and plasma/serum samples. We analyzed the ctDNA of patients, with a median follow-up of 357 days, and monitored CEA and CA19-9, and radiological imaging. Personalized mutational profiles derived from tumor tissue via whole-exome sequencing were used to design patient-specific ctDNA assays for variant detection in plasma samples (Signatera test, Natera).
Results: We found that during the course of the follow-up, 44.4% (4/9) of patients relapsed. The presence of ctDNA was associated with reduced recurrence-free survival (p = 0.011). Importantly, ctDNA results were found to correlate and precede imaging results. In contrast, CA 19-9 and CEA, in certain cases showed discordance with imaging and were found to be elevated due to other benign conditions, such as gastritis.
Conclusion: This study demonstrates that presence of ctDNA after surgery in early-stage PDAC is associated with reduced recurrence-free survival. During monitoring, ctDNA was found to be a better prognostic marker compared to CA-19-9 and CEA and can be used to inform on disease status prior to imaging.
Keywords
<p>Pancreatic adenocarcinoma, Circulating tumor DNA, Carcinoembryonic antigen, Cancer antigen 19-9, Neoadjuvant chemotherapy, Tumor markers, Positron emission tomography/computed tomography</p>
Article Details
Simple Summary
This study aimed to evaluate and validate the relationship between circulating tumor DNA (ctDNA) and recurrence in pancreatic adenocarcinoma while evaluating the relationship between tumor markers (CEA and CA19-9) and ctDNA. Furthermore, the study evaluated molecular residual disease clearance as assessed by ctDNA in the neoadjuvant and adjuvant therapy settings for pancreatic adenocarcinoma patients.
Nine patients with pancreatic cancer were enrolled (8 patients with pancreatic cancer and 1 patient with ampullary adenocarcinoma). The ctDNA samples were collected during the patients’ follow-up visits, with a median of 357 days (interquartile range: 248-542 days). Additional monitoring included carcinoembryonic antigen (CEA), cancer antigen 19-9 (CA 19-9), and radiological imaging. Personalized mutational profiles derived from tumor tissue via whole-exome sequencing were used to design patient-specific ctDNA assays for variant detection in plasma samples (Signatera test, Natera). Our findings suggest that the presence of ctDNA after surgery in early-stage PDAC is associated with reduced recurrence-free survival. During monitoring, ctDNA was found to be a better prognostic marker compared to CA-19 9 and CEA and it can be used to inform on disease status prior to imaging.
Introduction
Pancreatic adenocarcinoma (PDAC) has one of the most aggressive cancer histologies, with high recurrence (85%) and a 5-year survival rate of 9% [1,2]. Frequently, at the time of diagnosis curative surgery is not a treatment option. For these cases of locally advanced or metastatic pancreatic cancer, Fluorouracil-based and gemcitabine-based chemotherapy is the usual treatment and despite significant improvements in combination treatment for pancreatic cancer, the 5-year survival rate for these patients is only 5% [2,3]. CA19-9 levels are commonly utilized in clinical settings to predict prognosis and to evaluate tumor response to pancreatic cancer treatments, but CA19-9 levels may be abnormally high in patients with benign non-malignant conditions like biliary obstruction or infection and may be undetectable in Lewis antigen-negative individuals.
Circulating tumor cells (CTCs), which are tumor cells shed from primary or metastatic locations that enter the peripheral circulation, have recently emerged as a potential biomarker. Cell-free DNA was first discovered in blood plasma in 1948 by Mandel and Metais [4]. In 1965 and 1966, it was also found that cell -free DNA could be involved in metastasis and associated with disease. In fact, elevated levels of cell-free DNA were found in patients with systemic lupus erythematosus [5,6]. About 30 years after it was first discovered, Leon and colleagues utilized radioimmunochemistry to demonstrate that there was an elevation of cell-free DNA in cancer patients compared to normal control subjects [7]. However, due to technological limitations, it was not until 1994 that cell-free DNA was discovered to be directly originated from tumor cells [8,9]. In 1994, two studies found tumor-specific mutations in cell-free DNA of plasma samples from patients with pancreatic adenocarcinoma and acute myelogenous leukemia. In these studies, mutation-specific primers were used for polymerase chain reaction (PCR) amplification of N-RAS mutations. CtDNA, which is typically derived from CTCs and/or primary tumor cells, is usually diluted by normal DNA in bodily fluids. As a result, the existing sequencing approaches in the 90s, such as Sanger sequencing, were not sufficiently sensitive at detecting mutant ctDNA molecules. Mutation-specific PCR approaches were the only available technology that allowed better detection of the weak tumor-specific cell-free DNA signal. This specific approach became the preferred method to assess mutation status in cell-free DNA until the rise of massive parallel sequencing/next generation sequencing technology [10]. From 1997-2000, cell-free DNA started to be clinically implemented in prenatal diagnosis of sex determination and pregnancy-associated disorders by assaying fetal DNA in maternal plasma [11-13]. In 2005, a digital PCR method known as beads, emulsion, amplification, magnetic was utilized to detect and quantify mutations in the plasma of patients with colorectal tumors. [14]. This study found that patients with advanced colorectal cancers consistently had mutant adenomatous polyposis coli (APC) DNA molecules in their plasma. In >60% of patients with early and potentially curable colorectal cancer, mutant APC DNA molecules were detected at levels ranging from 0.01 to 1.7% of the total APC molecules, suggesting that tumoral DNA released in the circulation can have diagnostic purpose. Since the advancement of digital PCR and NGS-based technologies to detect ctDNA, ctDNA has become a biomarker in multiple cancer types, including lymphoma, thyroid cancer, breast cancer, gastrointestinal stromal tumors, colorectal cancer, and lung cancer but not pancreatic cancer yet and it particularly useful to detect common mutations in genes such as BRAF, EGFR, PIK3CA, KRAS, P53, ESR1, PDGFRA, etc.). [15-24]. Due to these promising earlier studies, ctDNA has now been utilized in clinical trials, particularly because ctDNA analysis will allow clinical investigators to monitor tumors in response to targeted therapy, monitor development of resistance, and detect minimal residual disease [25-27]. Recently, numerous circulating biomarkers have been evaluated for the detection of molecular residual disease (MRD) and molecular monitoring in PDAC. More recent studies have demonstrated the usefulness of ctDNA in minimal residual disease surveillance in HCC [28-30]. For example, Kasi et al, analyzed 200 plasma samples from a total of 90 hepatobiliary patients, with 27 of these patients having HCC [31]. It was reported that ctDNA detection was significantly associated with the stage of disease. In addition, serial time point analysis was performed on a subset of patients (n=56) that had 2-7 time points available and correlations between ctDNA levels and clinical response were noted and presented [29,31,32]. However, the application of these techniques has been limited in early-stage PDAC due to poor sensitivity and specificity [2]. More recently, an ultrasensitive, personalized, and tumor-informed ctDNA assay has shown to overcome many of the challenges that have plagued the aforementioned biomarkers, thus allowing for reliable detection of MRD across different tumors.
Materials and Methods
This single center study retrospectively reviewed patients with pancreatic cancer at the Houston Methodist Cancer Center who have data available for ctDNA testing. A total of nine metastatic pancreatic cancer patients were retrospectively reviewed for ctDNA analysis between May 2019 and March 2021. Revewing patients charts, all plasma samples were collected from 8 patients with pancreatic cancer and 1 patient with ampullary adenocarcinoma postoperatively prior to and after adjuvant therapy per patients follow up office visits and surveillance scans.
For all patients, data on post-surgery clinical intervention and other clinicopathologic information were obtained and followed up for a median of 357 days (interquartile range: 248-542 days). In order to confirm the diagnosis, the pathology of these patients was determined using biopsy samples obtained by the oncological surgeons of Houston Methodist Cancer Center. All patients were treated and followed up in accordance with The National Comprehensive Cancer Network (NCCN) recommendations.
Personalized mutational profiles derived from tumor tissue via whole-exome sequencing were used to design patient-specific ctDNA assays for variant detection in plasma samples. Multiplex PCR primer pairs for the selected set of variations were created. Cell-free DNA was isolated from a median of 8.5 mL of plasma (interquartile range, 7.5-9-5 mL). As previously reported, A-tailing, and ligation using an adapter were used to generate universal libraries, and following that, libraries were amplified using multiplex PCR, barcoded, pooled, and sequenced on an NGS sequencing platform (HiSeq2500 system, Illumina Inc). ctDNA plasma samples were classified as positive if they had at least two variants identified per provider (Signatera test, Natera).
Apart from ctDNA analysis, patients were also monitored using carcinoembryonic antigen (CEA), cancer antigen 19-9 (CA 19-9), and radiological imaging. In addition, computed tomography (CT) or Magnetic resonance imaging (MRI) was used to evaluate the treatment response every 8-12 weeks according to the guidelines of Response Evaluation Criteria in Solid Tumors (RECIST) 1.0.
Recurrence-free survival (RFS) was the major outcome measure, as determined by conventional radiologic criteria. Recurrence-free survival was calculated from the date of surgery to the first confirmed radiologic recurrence (local or distant) or death due to pancreatic cancer and was censored at the final follow-up or non-pancreatic cancer- related death. The Kaplan-Meier analysis was performed between ctDNA positive and negative groups, and between CA19-9 elevated and normal groups. Log-rank test was used to test the overall survival between the two groups. Statistical significance was defined as two-tailed p<0.05 for all tests. Stata IC/12.1 (StataCorp) and R statistical software, version 2.4 for Windows, were used to conduct the statistical analysis (R Foundation for Statistical Computing). Natera Inc performed the ctDNA analysis retrospectively, with scientists blinded to patient outcome and sample order.
|
Patient ID |
ctDNA Status |
Sex |
Age |
CA 19-9 Status |
Surgery Date |
Relapse |
Date of Recurrence or Last follow up visit |
|
1 |
Negative |
F |
63 |
Elevated |
4/17/2019 |
No |
7/15/2021 Last follow up |
|
2 |
Positive |
F |
77 |
Elevated |
02-12-2020 |
Yes |
11/17/20 Recurrence |
|
3 |
Positive |
M |
42 |
Elevated |
06-06-2019 |
Yes |
11/29/20 Recurrence |
|
4 |
positive |
F |
49 |
Elevated |
09-02-2020 |
Yes |
2/9/21 Recurrence |
|
5 |
Negative |
F |
70 |
Normal |
03-11-2020 |
No |
7/15/2021 Last follow up |
|
6 |
Negative |
M |
63 |
Normal |
09-10-2019 |
No |
7/22/2021 Last follow up |
|
7 |
Positive |
F |
59 |
Elevated |
05-05-2020 |
Yes |
1/8/21 Recurrence |
|
8 |
Negative |
M |
71 |
Normal |
7/14/2020 |
No |
7/6/2021 Last follow up |
|
9 |
Negative |
M |
83 |
Normal |
01-05-2021 |
No |
7/1/2021 Last follow up |
Table 1: ctDNA: Circulating tumor DNA; CA19-9: Cancer antigen 19-9. Patinates highlighted with in green had negative ctDNA that is associated with no recurrences of cancer, while all the patinates highlighted in red had positive ctDNA test that is associated with disease relapse.CA 19-9 was elevated in patient#1 however, no recurrence was reported.
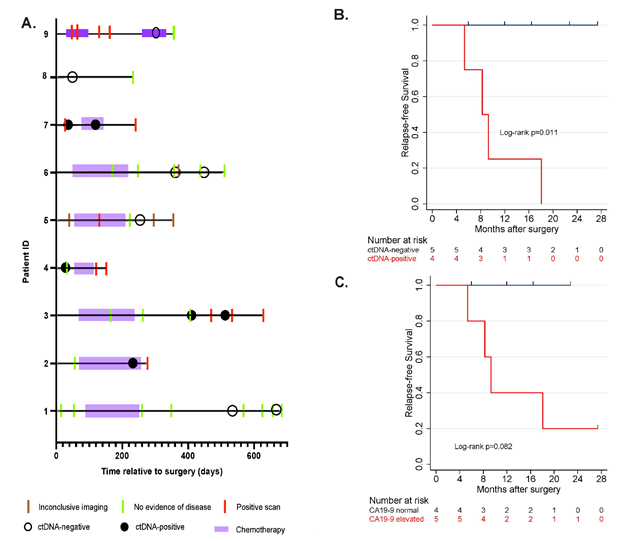
Figure 1: A. Patient over view plot, B,C. association of relapse- free survival with ctDNA and CA-19-9 status in longitudinal setting
1A: In this cohort, post- surgical ctDNA analysis detected relapse in 4 patients with a median follow up of 357 days (interquartile range: 248-542 days). B,C: Present the estimated relapse- free survival distributions by ctDNA and CA 19-9. The result of log rank test indicates significant differences between the survival curves between patints with ctDNA positive with ctDNA negative (p=0.011). the survival curves between patients with CA 19-9 elevated and CA 19-9 normal were not significant different (p=0.082). Moreover, CA 19-9 and CEA showed discordance with imaging and were elevated due to other conditions, such as gastritis.
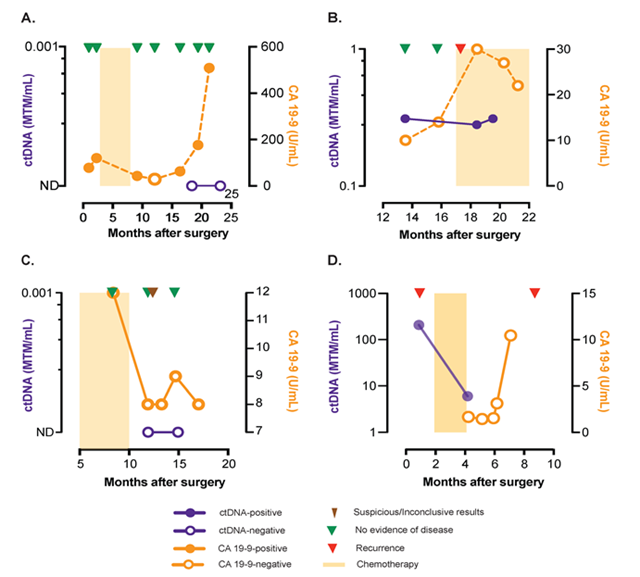
Figure 2: ctDNA for early relapse detection and disease status monitoring
Figure 2A-D: During monitoring, ctDNA is a better prognostic marker compared to CA-19-9.
B,D: Three patients had collected plasma after surgery, of which 2 patients (MRD rate 66%) were found to be ctDNA-positive with both experiencing relapse. MTM: Mean tumor molecules.
Results
Circulating tumor DNA associated with minimal residual disease and relapse
In this retrospective study, post-surgical ctDNA analysis detected relapse in 4 patients with a median follow-up of 357 days (interquartile range: 248-542 days; Figure 1A). During follow-up, 44.4% (4/9) of patients relapsed and of these patients, 100% had ctDNA detected prior to or at the time of recurrence (100% sensitivity and specificity. Presence of ctDNA (Figure 1B) was significantly associated with reduced relapse-free survival (log-rank p=0.011). In comparison, CA 19-9 (Figure 1C) showed inferior performance in predicting relapse. The survival curves between patients with CA19-9 elevated and CA19-9 normal were not significant different (p=0.082). Moreover, CA-19-9 and CEA showed discordance with imaging and were elevated due to other conditions, such as gastritis. During monitoring, ctDNA was a better prognostic marker compared to CA-19-9 (Figure 2A - D). Three patients had plasma collected after surgery and 2 of these patients (Minimal residual disease MRD rate of 66%) were found to be ctDNA-positive with both patients experiencing relapse (Figure 2B, D).
Table 1 showed that all patinates with negative ctDNA results (highlighted in green) showed no recurrences even with falsely elevated Ca19-9 (patient #1), while patinates with positive ctDNA values (highlighted in red) showed disease recurrence.
Circulating tumor DNA associated with disease status: Patients’ presentations
Figure 3 highlights patients’ presentations and demonstrates that ctDNA test result can be associated with disease status more accurately that representative tumor markers.
In patient number 1, who was diagnosed with pancreatic cancer in April 2019 and then had surgery in the same month, surveillance post operation CT scans revealed no evidence of disease (NED). Adjuvant chemotherapy (ACT) was started in July 2019 and continued until December 2019. CT was performed several times during 2020 and showed NED. Similarly, ctDNA analysis done in October 2020 showed a negative value. During the same time period, in October 2020, CA19-9 was detected at a high level but following EDG the CA19-9 elevation was linked to gastritis. These results suggest that ctDNA may be a better prognostic marker than CA-19-9 for pancreatic cancer. In patient 5, ctDNA was negative in November 2020 and the subsequent CT scans showed NED around the same time. Similarly, in patient 6, the value of ctDNA was connected to the disease course where ctDNA revealed a negative value in September and December 2020 and CT scans indicated NED during the same time period. For Patient 7, who had Whipple surgery in May 2020, CT scans performed in June 2020 showed the presence of residual disease. At the same time, ctDNA analysis matched the diagnosis. Patient 9 was diagnosed with pancreatic cancer in June 2020, received FOLFIRINOX from June 2020 to September 2020. The imaging was repeated in October 2020, confirming persistence of disease ctDNA was negative in April 2021 and subsequent CT scan performed in May 2021 was consistent with this result (Figure 3).
Circulating tumor DNA informs prior to surveillance imaging:
Regarding patient 2, who diagnosed in January 2020 and had Whipple surgery in February 2020 the CT revealed NED on April 2020. The patient started to receive mFOLFIRINOX and was then switched to Gemcitabine/ oxaliplatin in July 2020.The ctDNA showed positive results in October 2020 prior to the CT done in November 2020 that informed of recurrence (Figure 4).
Interestingly, in-patient number 3, who had Whipple surgery in June 2019, imaging showed NED until July 2020, whereas ctDNA analysis performed at the same time, revealed a positive value indicative of recurrence. At a later time, in November 2020, CT confirmed the recurrence demonstrating that ctDNA was informing us of recurrence in advance to imaging., Since pancreatic cancer is an extremely aggressive disease, early markers of disease progression are of great value for appropriate treatment decisions (Figure 4). The ctDNA of Patient 4 detected the recurrence in September 2020 while the CT scan showed NED until January 2021. After that date, the CT confirmed the recurrence. These findings suggest that ctDNA can detect earlier signs of illness that could aid in critical treatment decisions Patient 8 had a pancreatic mass diagnosed in June 2020, and then Whipple surgery in July 2020. In September 2020, ctDNA revealed negative results prior to the CT scan performed in March 2021 that confirmed NED (Figure 4).
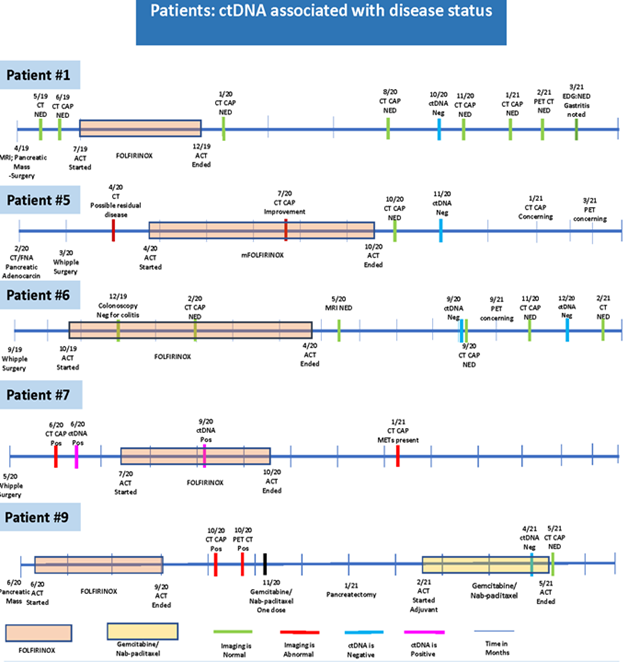
Figure 3: CT: computerized tomography; CAP: chest, abdomen and pelvis; MRI: magnetic resonance imaging; PET: positron emission tomograohy; EUS: endoscopic ultra sound; ERCP: endoscopic retrograde cholangiopancreatography; NED: no evidence of disease; act: a chemotherapy; ctDNA: circulating tumor DNA; EDG: esophagogastroduodenoscopy; Neg: negative; Pos: positive; METs: metastasis; FNA: fine needle aspiration
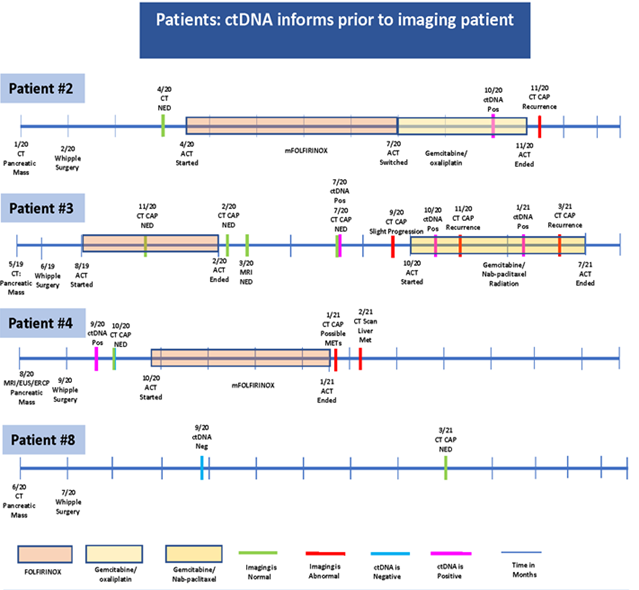
Figure 4: CT: computerized tomography; CAP: chest, abdomen and pelvis; MRI: magnetic resonance imaging; PET: positron emission tomograohy; EUS: endoscopic ultra sound; ERCP: endoscopic retrograde cholangiopancreatography; NED: no evidence of disease; act: a chemotherapy; ctDNA: circulating tumor DNA; EDG: esophagogastroduodenoscopy; Neg: negative; Pos: positive; METs: metastasis; FNA: fine needle aspiration
Discussion
The analysis of ctDNA represents a unique method to explore the DNA in the bloodstream that comes from cancerous cells and tumors the clinical arena. To our knowledge, this is one of the first retrospective study to investigate the validity of using ctDNA test in pancreatic cancer patients in correlation with tumor markers such as CA19-9 and imaging. One of the first stages in establishing a cancer diagnosis is to do a complete molecular examination of tissue biopsy samples. However, due to the difficulties in collecting sufficient tissue samples in patients with pancreatic cancer, the clinical value of such a study is typically decreased. As a result, there has been a lot of attention on developing and implementing highly accurate and less invasive cancer monitoring blood tests. Although various serological biomarkers are utilized in the clinic for pancreatic patients, such as CEA and CA 19–9, they are neither sensitive nor specific enough for prognostication. It has shown that ctDNA is produced from the original tumor and/or its metastases, and that it can be detected using next-generation sequencing or ddPCR with great sensitivity(33, 34). As a result, it might be utilized for cancer surveillance and minimal residual disease evaluation.
The ctDNA biopsy offers a noninvasive approach that potentially resolves the limited access to pancreatic cancer tissue samples by tissue biopsy. In addition, ctDNA biopsy reveals a dynamic picture of pancreatic cancer disease status, easily repeatable when needed, and provides real-time surveillance for minimal residual disease and long-term monitoring in pancreatic cancer patients that is more informative than the current tumor markers.
In our study, a total of nine patients with resectable pancreatic or ampullary adenocarcinoma who had ctDNA data available were retrospectively analyzed. The mean age was 64.1 ± 13.0 years and the male population represented 44.4 % (n=4). All plasma samples from 9 patients with a median follow-up of 357 days (interquartile range: 248-542 days) were analyzed and quantified using ultradeep multiplex PCR–based NGS per provider (Signatera test, Natera).
Eight patients received mFOLFIRINOX adjuvant chemotherapy, two of them received additional rounds of gemcitabine with Nab-paclitaxel and one received gemcitabine with oxaliplatin. Three patients had plasma collected after surgery and 2 of these patients were found to be ctDNA-positive and eventually relapsed. During the course of the follow-up, 44.4% (4/9) of patients relapsed (Table 1). The presence of ctDNA was associated with reduced recurrence-free survival (p = 0.011). Circulating tumor DNA findings were found to correlate and precede imaging results (Figure 4). In contrast, CA 19-9 and CEA levels, in certain cases, showed discordance with imaging results and were found to be elevated due to other benign conditions, such as gastritis.
We found during monitoring, ctDNA is a better prognostic marker compared to CA-19-9 (Figure 2A-D). Three patients had plasma collected after surgery, of which 2 patients (MRD rate of 66%) were found to be ctDNA-positive with both experiencing relapse. (Figure 2B, D)
While this stay is a retrospective study on small number of patients, it suggests and highlights the clinical utility of ctDNA in an aggressive GI cancer like pancreatic adenocarcinoma that is known for short recurrence. A cohort study in manner of prospective follow-up and successive blood collection may be necessary in the future to demonstrate the efficacy of using ctDNA as biomarker for pancreatic cancer MRD and surveillance which is our next planned prospective study.
Conclusions
Our findings suggest, presence of ctDNA after surgery in early-stage PDAC is associated with reduced recurrence-free survival. During monitoring, ctDNA was found to be a better prognostic marker compared to CA-19 9 and CEA and can be used to inform on disease status prior to imaging. More studies are needed to evaluate using ctDNA as biomarker for pancreatic cancer MRD and surveillance which is our next planned prospective study.
Author Contributions
Abdelrahim M, Esmail A, contributed to the conception; Abdelrahim M , Esmail A and Malhotra M contributed to abstract writing; Abdelrahim M, Esmail A, contributed to literature search and acquisition; Abdelrahim M, Esmail A are responsible for drafting and revising the manuscript; Abdelrahim M, Esmail A, Xu J, Tiffany A. Katz T, Sharma S, Kalashnikova E, Malhotra M, Olshan P, Billings P and Aleshin A contributed to analysis and interpretation; Abdelrahim M was the patients oncologist and was responsible for the critical revision of the manuscript for intellectual content; and all authors issued final approval for the version to be submitted. All authors affirm final approval of the version to be published and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Acknowledgments and Disclosures
M.A., A.E. and J.X. are employees of Houston Methodist Cancer Center, Houston, Tx. All other authors are employees of Natera, Inc. with stocks or options to own stock in the company. This article development support was provided by Minu Maninder, Ph.D. from Natera, Inc.
Conflicts of interest/Financial disclosures
None reported. All authors have declared there are no financial conflicts of interest with regard to this work.
References
- Society AC. Cancer Facts & Figures (2021).
- Loft M, Lee B, Tie J, et al. Clinical applications of circulating tumour DNA in pancreatic adenocarcinoma. J Pers Med 9 (2019).
- Lee JC, Ahn S, Cho IK, et al. Management of recurrent pancreatic cancer after surgical resection: a protocol for systematic review, evidence mapping and meta-analysis. BMJ Open 8 (2018): e017249.
- Mandel P, Metais P. [Nuclear Acids In Human Blood Plasma]. C R Seances Soc Biol Fil 142 (1948): 241-243.
- Tan EM, Schur PH, Carr RI, et al. Deoxybonucleic acid (DNA) and antibodies to DNA in the serum of patients with systemic lupus erythematosus. J Clin Invest 45 (1966): 1732-1740.
- Bendich A, Wilczok T, Borenfreund E. Circulating DNA as a possible factor in oncogenesis. Science 148 (1965): 374-376.
- Leon SA, Shapiro B, Sklaroff DM, et al. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res 37 (1977): 646-650.
- Vasioukhin V, Anker P, Maurice P, et al. Point mutations of the N-ras gene in the blood plasma DNA of patients with myelodysplastic syndrome or acute myelogenous leukaemia. Br J Haematol 86 (1994): 774-779.
- Sorenson GD, Pribish DM, Valone FH, et al. Soluble normal and mutated DNA sequences from single-copy genes in human blood. Cancer Epidemiol Biomarkers Prev 3 (1994): 67-71.
- Volik S, Alcaide M, Morin RD, et al. Cell-free DNA (cfDNA): Clinical significance and utility in cancer shaped by emerging technologies. Mol Cancer Res 14 (2016): 898-908.
- Lo YM. Fetal DNA in maternal plasma: biology and diagnostic applications. Clin Chem 46 (2000): 1903-1906.
- Lo YM, Corbetta N, Chamberlain PF, et al. Presence of fetal DNA in maternal plasma and serum. Lancet 350 (1997): 485-487.
- Lo YM, Tein MS, Lau TK, et al. Quantitative analysis of fetal DNA in maternal plasma and serum: implications for noninvasive prenatal diagnosis. Am J Hum Genet 62 (1998): 768-775.
- Diehl F, Li M, Dressman D, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci USA 102 (2005): 16368-16373.
- Gonzalez-Cao M, Mayo-de-Las-Casas C, Molina-Vila MA, et al. BRAF mutation analysis in circulating free tumor DNA of melanoma patients treated with BRAF inhibitors. Melanoma Res 25 (2015): 486-495.
- Kang G, Bae BN, Sohn BS, et al. Detection of KIT and PDGFRA mutations in the plasma of patients with gastrointestinal stromal tumor. Target Oncol 10 (2015): 597-601.
- Lubitz CC, Parangi S, Holm TM, et al. Detection of Circulating BRAF(V600E) in patients with papillary thyroid carcinoma. J Mol Diagn 18 (2016): 100-108.
- Piotrowska Z, Niederst MJ, Karlovich CA, et al. Heterogeneity underlies the emergence of EGFRT790 wild-type clones following treatment of T790M-positive cancers with a third-generation EGFR Inhibitor. Cancer Discov 5 (2015): 713-722.
- Roschewski M, Dunleavy K, Pittaluga S, et al. Circulating tumour DNA and CT monitoring in patients with untreated diffuse large B-cell lymphoma: a correlative biomarker study. Lancet Oncol 16 (2015): 541-549.
- Schiavon G, Hrebien S, Garcia-Murillas I, et al. Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci Transl Med 7 (2015): 313ra182.
- Sefrioui D, Perdrix A, Sarafan-Vasseur N, et al. Short report: Monitoring ESR1 mutations by circulating tumor DNA in aromatase inhibitor resistant metastatic breast cancer. Int J Cancer 137 (2015): 2513-2519.
- Spindler KL, Pallisgaard N, Andersen RF, et al. Circulating free DNA as biomarker and source for mutation detection in metastatic colorectal cancer. Plos One 10 (2015): e0108247.
- Xu S, Lou F, Wu Y, et al. Circulating tumor DNA identified by targeted sequencing in advanced-stage non-small cell lung cancer patients. Cancer Lett 370 (2016): 324-331.
- Yoo C, Ryu MH, Na YS, et al. Analysis of serum protein biomarkers, circulating tumor DNA, and dovitinib activity in patients with tyrosine kinase inhibitor-refractory gastrointestinal stromal tumors. Ann Oncol 25 (2014): 2272-2277.
- Arnedos M, Vicier C, Loi S, et al. Precision medicine for metastatic breast cancer--limitations and solutions. Nat Rev Clin Oncol 12 (2015): 693-704.
- Polivka Jr., Pesta M, Janku F. Testing for oncogenic molecular aberrations in cell-free DNA-based liquid biopsies in the clinic: are we there yet? Expert Rev Mol Diagn 15 (2015): 1631-1644.
- Siravegna G, Mussolin B, Buscarino M, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med 21 (2015): 795-801.
- Cai Z, Chen G, Zeng Y, et al. Comprehensive liquid profiling of circulating tumor DNA and protein biomarkers in long-term follow-up patients with hepatocellular carcinoma. Clin Cancer Res 25 (2019): 5284-5294.
- Jiang P, Chan CW, Chan KC, et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc Natl Acad Sci 112 (2015): E1317-E1325.
- Ono A, Fujimoto A, Yamamoto Y, et al. Circulating Tumor DNA Analysis for Liver Cancers and Its Usefulness as a Liquid Biopsy. Cell Mol Gastroenterol Hepatol 1 (2015): 516-534.
- Kasi PM, Budde G, Dayyani F, et al. Tumor-informed assessment of circulating tumor DNA and its incorporation into practice for patients with hepatobiliary cancers. Journal of Clinical Oncology 39 (2021): 4103-4109.
- Labgaa I, Villacorta-Martin C, D'Avola D, et al. A pilot study of ultra-deep targeted sequencing of plasma DNA identifies driver mutations in hepatocellular carcinoma. Oncogene 37 (2018): 3740-3752.
- Pietrasz D, Pecuchet N, Garlan F, et al. Plasma Circulating Tumor DNA in Pancreatic Cancer Patients Is a Prognostic Marker. Clin Cancer Res 23 (2017): 116-123.
- Alix-Panabières C, Pantel K. Circulating tumor cells: liquid biopsy of cancer. Clin Chem 59 (2013): 110-118.


 Impact Factor: * 4.2
Impact Factor: * 4.2 Acceptance Rate: 72.62%
Acceptance Rate: 72.62%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 