A Rare Case of Periprosthetic Streptobacillosis - Rapid Identification via Nanopore Sequencing after Inconclusive VITEK MS Results
Riccardo Spott1,#,*, Benjamin T Schleenvoigt¹,#, Birgit Edel2, Mathias W Pletz¹, Christian Brandt1
¹Institute for Infectious Diseases and Infection Control, Jena University Hospital, Jena, Germany
²Institute for Medical Microbiology, Jena University Hospital, Jena, Germany
# Both authors contributed equally to this work
*Corresponding Author: Riccardo Spott, Institute for Infectious Diseases and Infection Control, Jena University Hospital, Jena, Germany.
Received: 07 July 2022; Accepted: 20 July 2022; Published: 29 August 2022
Article Information
Citation: Riccardo Spott, Benjamin T Schleenvoigt, Birgit Edel, Mathias W Pletz, Christian Brandt A Rare Case of Periprosthetic Streptobacillosis - Rapid Identification via Nanopore Sequencing after Inconclusive VITEK MS Results. Archives of Clinical and Medical Case Reports 6 (2022): 513-517.
View / Download Pdf Share at FacebookAbstract
Streptobacillus moniliformis is the main cause of human streptobacillosis, also known as rat-bite or Haverhill fever. A correct diagnosis might be delayed due to scarcity, non-specific symptoms, and difficulties in cultivating this fastidious organism. We report on an 85-year-old man with periprosthetic streptobacillosis, which manifested itself in the form of immobilizing hip pain in the left prosthetic hip joint. The patient denied fever, chills, and night sweats. No fraction of the left hip could be detected via x-ray. Punctuatio.n of the hip joint revealed putrid aspirate and computer tomography detected an abscess in the left hip. From the synovial fluid and from concomitantly drawn blood cultures grew a pleomorphic, gramnegative rod after 48 h and 35 h, respectively. The VITEK and VITEK MS (MALDI-TOF) analysis remained inconclusive. Therefore, the unknown isolate was sequenced using Nanopore Third-Generation Sequencing. After one hour enough data were generated to identify the microorganism as Streptobacillus moniliformis.
To date, this is the first published case of identifying Streptobacillus moniliformis by rapid whole genome sequencing as the cause of a periprosthetic infection. Despite a mortality rate of around 10% in humans, streptobacillosis is rarely diagnosed and published. In this context, nanopore sequencing enabled fast and reliable diagnostic results for identifying microorganisms and their properties
Keywords
<p>Oxford Nanopore Sequencing; Streptobacillosis; <em>Streptobacillus moniliformis</em>; WGS; Zoonosis</p>
Article Details
1. Background
Streptobacillus moniliformis is the main cause of human streptobacillosis. Depending on the transmission, it is distinguished into “rat-bite”-fever (RBF) and “Haverhill“-fever (HF) [1]. RBF is transferred by bites or scratches of rats and other rodents, whereas HF-transmission occurs by rat-excrement polluted water and food. Symptoms are relatively unspecific, comprising more general symptoms like fever, chills, vomiting, malaise, muscle pain and rash [2,3]. They typically start to appear within the first 10 days of infection [3]. However, more serious complications such as arthritis, endocarditis and bacteremia can occur as the disease progresses [3]. Cultivation of Streptobacillus moniliformis proves to be challenging, with growth occurring only after 2-7 days under microaerophilic conditions [3,4]. Overgrowth by faster-growing bacteria in the specimen can therefore hinder the cultivation [4]. Due to the unspecific first symptoms and difficulties in the cultivation, wrong or delayed diagnosis is not unlikely [3].
2. Case Report
An 85-year-old man has been hospitalized with pain and immobilization of his left hip joint prosthesis for two days. The implantation was 26 years previously and no previous complications are mentioned in the report. He denied typical infection symptoms such as fever and shivering and the temperature on admission was not given in the available reports. X-ray examination revealed no fracture of the hip, but C - Reactive Protein (CRP) and Leukocytes were > 69.5 mg/l and 22.4 Gpt/l respectively. To rule out the suspicion of inflammation the hip joint was punctured, and putrid synovial fluid was collected and sent to the microbiology department for further analysis. Subsequent computer tomography revealed an abscess in the Musculus gluteus medius and the patient was diagnosed with a periprosthetic infection. Profound debridement, partial synovectomy and jet lavage of the left hip joint and muscular debridement of the left hip and thigh were performed. After this, an empirical treatment with Unacid intravenously was initialized. The replacement of the mobile prosthesis parts was not possible, because they are out of production. In the following discussion, to either fully replace the prosthesis or to try to preserve the actual one, the patient decided on the preservation of his actual prosthesis. The laboratory could detect an unknown microorganism after 35 hours of incubation in aerobic blood culture and after 48 hours in the puncture secretion. The colony morphology was described as small, round, convex gray colonies. Blood culture microscopy showed the microorganism as flagella-like, gram-negative rods (Figure 1 A). In contrast, microscopy of the culture specimen showed gram-negative, pleomorphic rods, partially swollen on their sides (Figure 1 B). Agar diffusion assays using Bestbion teststrips on MHF-agar (Oxoid) revealed that the unknown microorganism was sensitive to penicillin, amoxicillin/clavulanic acid, ceftriaxone, and tigecycline. VITEK2 and VITEK MS (MALDI-TOF; Server-version: Myla 4.8.0 / KB 3.2; VtkMS Plus 2.0 / Saramis 4.11 KB 4.16.0.; Knowledge base version 3.2) were used for species identification, but neither system could determine the species. Therefore, the microbiology department transferred the sample material to the Institute for Infectious Diseases and Infection Control for rapid whole-genome sequencing (WGS).
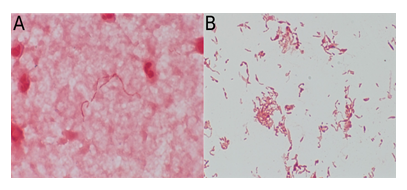
Figure 1: A) Flagella-like, gram-negative Streptobacillus moniliformis-rods in blood culture. B) Gram-negative, pleomorphic Streptobacillus moniliformis-rods in culture of the hip joint-aspirate.
The isolate could be reliably classified as Streptobacillus moniliformis 7.5 hours after the arrival of the material (DNA isolation 2.5 hours, sequencing library four hours, sequencing one hour). The Oxford Nanopore platform sequences in real time, meaning that during the run the so far produced data is available for analysis for e.g., bacterial identification. Unlike other DNA sequencing platforms, data from the Oxford Nanopore Platform is streamed in real-time and can be analyzed immediately, so there is no need to wait until the whole sequencing run is finished. Therefore, after approximately only one hour of sequencing, 10,000 raw DNA fragments (reads) were classified directly with Sourmash (Version 3.5.0) [5] using the GTDB database (Release 06-RS202 (April 27, 2021)) in less than 5 mins [6,7]. We reported the preliminary results directly to the microbiology department, and sequencing continued overnight. The subsequent complete reconstruction of the entire genome generated a circular 1.65 megabase large genome, which confirmed the preliminary results (the bacterial genome is available under the accession number: CP079714; 64489 total-reads: 41050 reads (63.5 %) assigned to Homo Sapiens, 23235 reads (36.03 %) assigned to Streptobacillis moniliformis). In agreement with the phenotypic antibiotic resistance testing, no resistance genes could be identified on the genome (genotype) using ABRicate (Version 1.0.1) [8] with the databases: CARD [9], NCBI AMRFinderPlus [10], PlasmidFinder [11], and Resfinder [12]. The initial Unacid treatment was therefore continued for a total of 14 days followed by an oral sequential therapy with amoxicillin/clavulanic acid (875 mg/ 125 mg; 1-1-1) for another 12 weeks. After 32 days of treatment, CRP had declined to > 6.6 mg/l and limited mobility was restored. The patient was released after 36 days in stabilized, still reduced, general condition upon his own request. A nursing service was engaged to take care of the antibiotic administration, but a follow-up report is not available. When asked specifically, he denied the keeping of domestic rats or rat bites and there was no evidence of outdoor activities with fresh water contacts in the patient's history.
3. Discussion
Streptobacillosis is rarely diagnosed [13,14], even though 10-100 % of domestic and 50-100 % of wild rats are colonized with Streptobacillus moniliformis [3]. The risk of infection by bite is assumed to be around 10 % [3], but other rodents or excrement-polluted food or water can also cause the infection [1]. If left untreated, the disease has a mortality rate of approximately 10 % [3]. Periprosthetic bone infection caused by Streptobacillus moniliformis was first reported in 2020 by Smallbones et al., describing a case of infection in a knee replacement [15]. A second publication by Fokkema et al. in 2022 also reported a case of periprosthetic joint infection, where S. moniliformis infected a knee arthroplasty [16]. In both cases 16S rRNA PCR was used for the identification of the microorganism. Therefore this publication is the third reported case with a periprosthetic focus in worldwide literature and the first utilizing rapid whole genome sequencing to identify the causative bacterium.
We conducted literature research of published case reports via PubMed and found 256 Publications using “Streptobacillus moniliformis” as a search term (August 2021), excluding 139 non-case reports. We further excluded 18 publications as they lacked information or were not eligible for inclusion as a Streptobacillus moniliformis infection (see Supplementary Data for details). The remaining 99 publications contained information on the number of reported cases and the focus of the infection. All cases were confirmed by culture or PCR. Survival, gender, age, and country of origin were extracted from the references. Time of survival after infection was not consistently reported in the case histories. Hence, the following case fatality rates are not based on uniform periods but reflect a summarized estimation. 36.9 % were female (41 out of 111). The median age was 36. The overall case-fatality rate was 9.9 % (11 out of 111 cases; 93 not fatal; 7 unknown). Most cases were reported from the USA (33.3 %; 37 out of 111) followed by the UK (11.7 %; 13/111), Japan (7.2 %; 8/111), and France, Germany, and Spain (4.5 % each; 5/111). 57 publications with 64 cases (57.7 %; 64/111) referred to bloodstream infections with positive blood cultures. The mortality of bloodstream infection was 12.5 % (8/64). Fifteen publications with 17 cases (15.3 %; 17/111) reported endocarditis as the focus of infection. The case fatality rate was 35.2 % (6/17). In 12 (18.8 %; 12/64) cases with bloodstream infection, endocarditis was proven simultaneously. Those cases showed a mortality of 41.6 % (5/12). In 40 cases (36.0 %; 40/111) arthritis was reported (mortality 2.5 %; 1/40). Other foci as meningitis, CNS Infection, Osteomyelitis, Spondylodiscitis, Chorioamnionitis, epidural abscess, and abscesses of spleen, brain, skin, psoas, or the small pelvis were rare. Slow growth and specific growth conditions can be significant obstacles in the identification of Streptobacillus moniliformis. The slower metabolism may cause weaker fluorescent biochemical reactions in the VITEK system that hampers reliable species identification in general [17–19].
However, even the VITEK MS system might fail to provide an accurate, species-level identification ranging from 1.5 to 22.2 % in gram-negative Isolates depending on the reporting study [20,21]. The failure to identify a species can have multiple origins, like unspecific or conflicting mass spectra, or due to the rarity of the disease, not enough good database references. The most precise form of species identification remains complete genome sequencing for accurate typing and genotyping (e.g., resistance or virulence genes) [22]. Highly curated genome databases such as GTDB with over 250,000 different bacteria and archaea can reliably identify even close relatives of unknown bacteria. In addition, long-read sequencing technologies such as PacBio or Nanopore sequencing already generate information-rich raw data (based on the length of the DNA) to enable reliable species identification without genome reconstruction. This approach was used to identify the Streptobacillus moniliformis at the beginning of the sequencing run, reducing the time to results by half a day. Classical 16S sequencing, an older routine sequencing-technique for the identification of microorganisms, bears the problem that only one single gene is sequenced and results may vary based on the primer used, primer binding efficiency and PCR performance in general. Furthermore in a best case scenario PCR and Sanger sequencing will take at least 4-5 h, which excludes DNA isolation, PCR preparation and data analysis [23]. Therefore rapid bacterial sequencing via Oxford Nanopore provides more data in less time while simultaneously generating a whole genome sequence for further characterization afterwards. The current barriers to broader routine-based sequencing applications remain price per sample and the bioinformatics know-how required to analyze sequencing data. However, prices are getting cheaper, and raw data is getting more accurate. In addition, scientists have been working very actively in recent years on so-called workflows to simplify the downstream analysis of sequencing data and reduce the bioinformatics bottleneck in applied science [24–26].
3. Conclusion
Streptobacillus moniliformis is a slow-growing pathogen that confers the rarely diagnosed zoonotic disease streptobacillosis. The slow-growing nature of this organism makes it especially difficult to identify via VITEK and the VITEK-MS, the current standards in routine clinical microbiology. Utilizing internal sequencing solutions designed for rapid typing can provide reliable results for all known bacteria or archaea species within a day. Further, WGS-analysis represents a "one-fits-all" approach as sequencing preparation between microorganisms remains mostly the same while also providing genotyping besides species identification. Sequencing is slowly becoming an essential tool for the routine microbiology laboratory besides outbreak tracing. The price per sample is continuously getting cheaper, new miniature device options are available, and bioinformatic workflows that simplify the downstream analysis are broadly available to non-scientific personnel.
Author contributions
Sample collection, microbiological testing and imaging, B.E.; sequencing, R.S.; software, C.B.; bioinformatic analysis, R.S., C.B.; literature research, B.T.S.; writing first draft, R.S.; reviewing and editing manuscript, R.S., C.B., B.T.S. and M.W.P.; supervision, C.B.; project administration, C.B.; funding acquisition, M.W.P. All authors have read and agreed to the published version of the manuscript.
Conflicts of Interest
The authors have declared that no competing interests exist.
Funding
This work was supported by grants from the Federal Ministry of Education and Research, [grant number 13GW0423B and 13GW0457A].
Patient Consent Statement
The patient gave his written agreement to the publication of this work.
References
- Gaastra W, Boot R, Ho HTK, et al. Rat bite fever. Vet Microbiol 133 (2009): 211-228.
- Eisenberg T, Ewers C, Rau J, et al. Approved and novel strategies in diagnostics of rat bite fever and other Streptobacillus infections in humans and animals. Virulence 7 (2016): 630-648.
- Elliott SP. Rat bite fever and Streptobacillus moniliformis. Clin Microbiol Rev 20 (2007):13-22.
- Eisenberg T, Ewers C, Rau J, et al. Approved and novel strategies in diagnostics of rat bite fever and other Streptobacillus infections in humans and animals. Virulence 7 (2016): 630-648.
- Brown CT, Irber L. sourmash. A library for MinHash sketching of DNA. J Open Source Softw 1 (2016): 27.
- Parks DH, Chuvochina M, Waite DW, et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol 36 (2018): 996-1004.
- Chaumeil PA, Mussig AJ, Hugenholtz P, et al. A toolkit to classify genomes with the Genome Taxonomy Database. Hancock J, editor. Bioinformatics (2019): btz848.
- Abricate ST. Github.
- Jia B, Raphenya AR, Alcock B, et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 45 (2017): D566-D573.
- Feldgarden M, Brover V, Haft DH, et al. Validating the AMR Finder Tool and Resistance Gene Database by Using Antimicrobial Resistance Genotype-Phenotype Correlations in a Collection of Isolates. Antimicrob Agents Chemother 63 (2019): e00483-19.
- Carattoli A, Zankari E, García-Fernández A, et al. In Silico Detection and Typing of Plasmids using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob Agents Chemother 58 (2014): 3895-3903.
- Zankari E, Hasman H, Cosentino S, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67 (2012): 2640-2644.
- Matt U, Schmiedel J, Fawzy A, et al. Infection in a Young Immunocompetent Male Caused by Streptobacillus felis, a Putative Zoonotic Microorganism Transmitted by Cats. Clin Infect Dis 72 (2021): 1826-1829.
- Hagelskjaer L, Sørensen I, Randers E. Streptobacillus moniliformis infection: 2 cases and a literature review. Scand J Infect Dis 30 (1998): 309-311.
- Smallbones M, Monem M, Baganeanu M, et al. Near-fatal Periprosthetic Infection with Streptobacillus moniliformis: Case and Review. J Bone Jt Infect 5 (2020): 50-53.
- Fokkema AT, Kampschreur LM, Pirii LE, et al. Rat bite fever in a total knee arthroplasty: an unusual case of periprosthetic joint infection. Arthroplasty 4 (2022): 13.
- Automated antimicrobial susceptibility testing of slow-growing Pseudomonas aeruginosa strains in the presence of tetrazolium salt WST-1 (2021).
- Ling TKW, Liu ZK, Cheng AFB. Evaluation of the VITEK 2 System for Rapid Direct Identification and Susceptibility Testing of Gram-Negative Bacilli from Positive Blood Cultures. J Clin Microbiol 41 (2003): 4705-4707.
- Eisenberg T, Nicklas W, Mauder N, et al. Phenotypic and Genotypic Characteristics of Members of the Genus Streptobacillus. Janssen PJ, editor. Plos One 10 (2015): e0134312.
- Yoo IY, Han J, Ha SI, et al. Clinical performance of ASTA SepsiPrep kit in direct bacterial identification and antimicrobial susceptibility test using MicroIDSys Elite and VITEK-2 system. J Clin Lab Anal 35 (2021): e23744.
- Manji R, Bythrow M, Branda JA, et al. Multi-center evaluation of the VITEK® MS system for mass spectrometric identification of non-Enterobacteriaceae Gram-negative bacilli. Eur J Clin Microbiol Infect Dis off Publ Eur Soc Clin Microbiol 33 (2014): 337-346.
- Viehweger A, Marquet M, Hölzer M, et al. Adaptive nanopore sequencing on miniature flow cell detects extensive antimicrobial resistence (2021).
- Church DL, Cerutti L, Gürtler A, et al. Performance and Application of 16S rRNA Gene Cycle Sequencing for Routine Identification of Bacteria in the Clinical Microbiology Laboratory. Clin Microbiol Rev 33 (2020): e00053-19.
- Hodcroft EB, De Maio N, Lanfear R, et al. Want to track pandemic variants faster? Fix the bioinformatics bottleneck. Nature 591 (2021): 30-33.
- Ewels PA, Peltzer A, Fillinger S, et al. The nf-core framework for community-curated bioinformatics pipelines. Nat Biotechnol 38 (2020): 276-278.
- Brandt C, Krautwurst S, Spott R, et al. poreCov-An Easy to Use, Fast, and Robust Workflow for SARS-CoV-2 Genome Reconstruction via Nanopore Sequencing. Front Genet 12 (2021): 711437.


 Impact Factor: * 5.3
Impact Factor: * 5.3 Acceptance Rate: 75.63%
Acceptance Rate: 75.63%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 
