Viral Integration Analysis Reveals Likely Common Clonal Origin of Bilateral HPV16-Positive, p16-Positive Tonsil Tumors
Lisa M. Pinatti1,2, Heather M. Walline2, Thomas E. Carey2, Jens Peter Klussmann3, Christian U. Huebbers2,4*
1Cancer Biology Program, Program in the Biomedical Sciences, Rackham Graduate School, University of Michigan, Ann Arbor, MI, USA
2Department of Otolaryngology/Head and Neck Surgery, University of Michigan, Ann Arbor, MI, USA
3Center for Molecular Medicine Cologne (CMMC), University of Cologne, Cologne, Germany
4Jean-Uhrmacher-Institute for Otorhinolaryngological Research, University of Cologne, Cologne, Germany
*Corresponding Author: Dr. Christian U. Huebbers, Jean-Uhrmacher-Institute for Otorhinolaryngological Research, University of Cologne, Cologne, Germany
Received: 08 April 2020; Accepted: 15 May 2020; Published: 13 July 2020
Article Information
Citation:
Lisa M. Pinatti, Heather M. Walline, Thomas E. Carey, Jens Peter Klussmann, Christian U. Huebbers. Viral Integration Analysis Reveals Likely Common Clonal Origin of Bilateral HPV16-Positive, p16-Positive Tonsil Tumors. Archives of Clinical and Medical Case Reports 4 (2020): 680-696.
View / Download Pdf Share at FacebookAbstract
Infections with high-risk human papilloma viruses (HPV) are responsible for a significant number of oropharyngeal squamous cell carcinoma (OPSCC), with infection rates currently rising at epidemic rates in the western world. Synchronous bilateral HPV+ tumors of both tonsils are a very rare event whose understanding, however, could provide important insights into virus-driven tumor development and progression and whether such integration events are of clonal origin. In this study we analyzed a single case of a bilateral tonsillar p16+ HPV+OPSCC. The viral integration status of the various tumor samples was determined by integration-specific PCR methods and sequencing, which identified viral insertion sites and affected host genes. Integration events were further confirmed by transcript analysis. Analysis of the tumors revealed common viral integration events involving the CD36 gene, as well as a unique event in the LAMA3 gene which resulted in loss of LAMA3 exon one in both tissues that had lost the complex viral LAMA3 integration event. In addition, there were several integration events into intergenic regions. This suggests a common origin but individual evolution of the tumors, supporting the single-clone hypothesis of bilateral tumor development. This hypothesis is further supported by the fact that the two cellular genes LAMA3 and CD36 as targets of viral integration are involved in cell migration and ECM-receptor interactions, which provides a possible mechanism for clonal migration from one tonsil to another.
Keywords
<p>Viral cancers, HPV; Head and Neck Cancer; Tumor Clonality; Tumor Metastasis</p>
Article Details
Abbreviations:
HPV- human papillomavirus; HPV+OPSCC- HPV-positive oropharyngeal squamous cell carcinoma; HPV+HNSCC- HPV-positive head and neck squamous cell carcinoma; DIPS-PCR- detection of integrated papillomavirus sequences PCR; FFPE- formalin-fixed paraffin-embedded
1. Introduction
Mucosal human papillomaviruses (HPV) are small double-stranded DNA viruses that infect human mucosa in the oral and anogenital tract. Persistent infection of HPV may lead to a variety of diseases, including genital warts, precancers, and cancers [1]. HPV-associated cancers of the head and neck (HPV+HNSCC), particularly HPV+ oropharyngeal squamous cell carcinoma (HPV+OPSCC), is a growing public health concern; the incidence of this cancer has increased at an epidemic rate in the last few decades [2, 3]. This is in contrast to HPV-negative HNSCC, which is mainly attributed to smoking and has been decreasing in incidence due to public health efforts to decrease smoking rates [2]. As a whole, HNSCC accounts for 650,000 cancer cases and 330,000 deaths annually worldwide [4]. In the United States, three of four newly diagnosed OPSCCs are HPV-positive, and it is predicted that by 2020, the prevalence of HPV+OPSCC will be higher than that of cervical cancer [5, 6]. This increasing incidence is not restricted to the United States but is observed for many countries of the western world including Germany [7].
Persistent oral infection of HPV is a risk factor for the development of OPSCC, but the rates of oral infection in the general population are relatively low and clearance of infections is common [8-10]. It is unclear why some HPV infections are cleared while others persist, but smokers, males, and individuals with higher numbers of sexual partners are more likely to have a persistent oral HPV infection [11]. The tonsils are the most common site of HPV+OPSCC; it is hypothesized that the tonsils act as a reservoir for HPV and the thin epithelium may be more susceptible to infection [9].
HPV drives carcinogenesis through disruption of critical cell cycle regulators TP53 and RB by the viral oncoproteins E6 and E7. Recently, the role of viral integration into the human genome has been investigated as an additional mechanism of carcinogenesis [12]. Viral integration is reported to take place throughout the cellular genome, including both gene-poor and gene-rich regions, and preferentially at fragile sites and regions of microhomology with the viral genome [13, 14]. Integration into or near genes can lead to changes in gene expression through a variety of mechanisms and can also lead to the generation of viral-human fusion transcripts, the implications of which are still widely unclear [12-15].
Very few patients with tonsillar carcinoma present with bilateral HPV+ tumors, and the literature on this phenomenon is somewhat limited [16-20]. A recent retrospective study reported that in a cohort of Danish patients, only 3.3% had synchronous bilateral tonsil cancer [21]. There are multiple hypotheses on the mechanism of this event. One is the single-clone hypothesis, in which carcinoma develops in one tonsil and a clonal population migrates away from the tumor to the other tonsil. Another is that both tonsils develop independent carcinomas due to HPV infection at both sites.
Here we present an interesting case of a 60-year-old Catholic nun who developed synchronous bilateral tonsillar p16+ HPV+OPSCC. Analysis of the tumors revealed both common viral integration events, as well as unique events. This suggests a common origin but individual evolution of the tumors, supporting the single-clone hypothesis of bilateral tumor development. Viral integration occurred into cellular genes LAMA3 and CD36; both of these genes are involved in cell migration and ECM-receptor interactions, which provides a possible mechanism for clonal migration from one tonsil to another.
2. Materials and Methods
2.1 Tumor Specimens
Formalin-fixed, paraffin-embedded tissue blocks were received as follows: Tumor 1/ block 1B (left tonsil), Tumor 2/blocks 2A, 2B, and 2D (right tonsil). Slides were prepared from each block and the tissues were reviewed by a head and neck pathologist. There was no tumor in block 2B.
2.2 DNA/RNA Isolation
10mm sections were taken from FFPE tissue blocks and mounted on a slide. Each section was aligned to the prepared H&E slides to identify the tumor-rich areas, and tissue within the tumor area was microdissected using a scalpel. Following microdissection, DNA and RNA were extracted from the tissue. DNA was isolated using the Machery-Nagel NucleoSpin DNA FFPE kit according to the manufacturer’s protocol. Briefly, paraffin was dissolved with xylene, and the tissue was lysed with lysis buffer and Proteinase K overnight at 56° C. Following overnight digestion, DNA was de-crosslinked, loaded onto the NucleoSpin DNA columns, washed and then eluted in water. DNA concentration was measured using the QUBIT 2.0 Fluorometer.
RNA was extracted from tissue using the Roche High Pure RNA Paraffin Kit according to the manufacturer’s protocol. Briefly, paraffin was dissolved using heptane and methanol, and the tissue was lysed overnight at 56ºC with lysis buffer containing Proteinase K and 10% SDS. RNA was extracted using the supplied High Pure Filters and wash buffers, followed by DNase I treatment. RNA was eluted in Elution Buffer and the concentration was measured using the QUBIT 2.0 Fluorometer.
2.3 P16 Staining
Tumor tissue sections were stained for p16 using the Roche/Cintec p16 mouse monoclonal antibody (#805-4713).
2.4 HPV Testing
HPV types present in the tumors were identified using HPV PCR-MassArray as previously described [22]. In brief, this method detects and identifies 15 high-risk HPV subtypes (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, 68, and 73), 2 low-risk subtypes (6 and 11), and HPV90, considered to be a possible high-risk subtype. The test included interrogation of human GAPDH as a control for sample DNA quality and assay validity. Type-specific, multiplex, competitive PCR was performed to amplify the E6 region of HPV, followed by probe-specific single base extension to discriminate between naturally occurring HPV present in the sample and the synthetic competitors included in the reaction. Matrix-assisted laser desorption/ionization time of flight mass spectroscopy was used for separation of products on a matrix-loaded silicon chip array. Samples were run in quadruplicate with appropriate positive and negative controls.
2.5 Detection of Integrated Papillomavirus Sequences (DIPS-PCR)
DIPS-PCR was performed to identify the sites of HPV integration in the genome of the tumors, as previously described [23]. For each tumor, 0.75mg DNA was digested with one of two restriction enzymes, either TaqA1 or Sau3AI. Adapters complementary to the unique overhangs created by restriction digestion were annealed to digested DNA. Linear PCR was performed on each sample using 11 different viral primers to amplify viral fragments. Following linear PCR, exponential PCR using 11 nested viral primers and an adapter-specific primer was performed. All DIPS-PCR primer sequences are listed in Table 2. Products of the exponential PCR reactions were separated by gel electrophoresis (3% agarose gel). Bands were excised from the gel and were purified by Qiagen Qiaquick Gel Extraction Kit. Sanger sequencing of the isolated products was performed by the University of Michigan Sequencing Core, and the results were mapped using NCBI BLAST.
|
Sample |
Viral insertion (nt) |
Map |
Integration locus |
Database comparison |
|
Left tonsil (1B) |
2088 (E1) |
Unplaced |
Genomic scaffold |
NT_187433.1 |
|
3886 (E5) |
4p15 |
Intergenic |
NC_000004.12 |
|
|
5617 (L2) |
7q21 |
CD36 intron 6 |
NG_008192.1 |
|
|
Right tonsil (2A) |
3867 (E5) |
18q11 |
LAMA3 intron 1 – intron 68 fusion |
NG_007853.2 |
|
Right tonsil (2D) |
3213 (E2) |
4q28 |
Intergenic |
NC_000004.12 |
|
3854 (E5) |
7q21 |
CD36 intron 5 |
NG_008192.1 |
Table 1: Integration locations found within the three samples.
|
Primer ID |
1st PCR Primer Name |
Primer Sequence |
Nested PCR Primer Name |
Nested Primer Sequence |
Episomal Size |
|
D1 |
HPV16-E1a |
5'-ACGGGATGTAATG GATGGTTTTATG-3' |
2nd-HPV16-E1a |
5'-AGGGGATGCTATA TCAGATGACGAG-3' |
7.5 kb |
|
D2 |
HPV16-E1b |
5'-ATGTTACAGGT AGAAGGGCG-3' |
2nd-HPV16-E1b |
5'-AGTCAGTATAG TGGTGGAAGTG-3' |
7.1 kb |
|
D3 |
HPV16-E1c |
5'-ACGCCAGAATGGA TACAAAGACAAAC-3' |
2nd-HPV16-E1c |
5'-ATGGTACAATGG GCCTACGATAATG-3' |
6.5 kb |
|
D4 |
HPV16-E2a |
5'-ACCCGCATGA ACTTCCCATAC-3' |
2nd-HPV16-E2a |
5'-TCAACTTGAC CCTCTACCAC-3' |
2.7 kb |
|
D5 |
HPV16-E5a |
5'-AGAGGCTGCTGT TATCCACAATAG-3' |
2nd-HPV16-E5a |
5'-ATGTAGACACA GACAAAAGCAGC-3' |
3.0 kb |
|
D6 |
HPV16-L2a |
5'-GTACGCCTAGA GGTTAATGCTGG-3' |
2nd-HPV16-L2a |
5'-CCAAAAAGTC AGGATCTGGAGC-3' |
3.5 kb |
|
D7 |
HPV16-L1a |
5'-ATCCACACCT GCATTTGCTGC-3' |
2nd-HPV16-L1a |
5'-GCACTAGCATTT TCTGTGTCATCC-3' |
5.5 kb |
|
D8 |
HPV16-E2b |
5'-GTGGACATTACAA GACGTTAGCCTTG-3' |
2nd-HPV16-E2b |
5'-CATGGATATACA GTGGAAGTGCAG-3' |
5.4 kb |
|
D9 |
HPV16-E2c |
5'-CGTCTACATGG CATTGGACAGG-3' |
2nd-HPV16-E2c |
5'-GATAGTGAATG GCAACGTGACC-3' |
4.7 kb |
|
D10 |
HPV16-L2b |
5'-CCACTTTACAT GCAGCCTCACC-3' |
2nd-HPV16-L2b |
5'-CTGTACCCTCTAC ATCTTTATCAGG-3' |
3.0 kb |
|
D12 |
HPV16-E6a |
5'-GTATTGCTGTT CTAATGTTGTTCC-3' |
2nd-HPV16-E6a |
5'-GCAAAGTCATAT ACCTCACGTCG-3' |
7.7 kb |
Table 2: DIPS-PCR primer sequences and product size given episomal virus input. Nested PCR reverse adapter primer: 5’-GATGCTGACGACTGATACCGG-3’
2.6 Direct PCR
Primers were designed to amplify the viral/cellular regions of integration identified by DIPS-PCR (Table 3). PCR was performed with genomic DNA from each tumor sample as well as DNA from the DIPS linear PCR reactions in order to enrich for viral products. Amplicons were separated and visualized with gel electrophoresis and were confirmed by Sanger Sequencing of the excised and purified bands.
|
Target |
F primer sequence |
R primer sequence |
PCR Product size (bp) |
RT-PCR Product size (bp) |
|
LAMA3 Exon 1 |
5’-CATATCCCC GGCTGCGCTA-3’ |
5’-GCCAGGTTGAA GTAAGTCGGG-3’ |
280 |
NA |
|
LAMA3 Exon 2 |
5’-CATCCTGTCAC CAATGCCATC-3’ |
5’-CCAAGGTGAGG TTGACTCTGTT-3’ |
97 |
97 |
|
LAMA3 Intron1:68 junction – HPV16 E5 |
5’-TGTATTTTTAG ATAAAGATGCCGC-3’ |
5’-ATGTAGACACA GACAAAAGCAGC-3’ |
260 |
260 |
|
CD36 Exon 4 |
5’-TGGGTTAAA ACAGGCACAGAA-3’ |
5’-ACTTGAATGTTGCT GCTGTTCA-3’ |
95 |
NA |
|
CD36 Exon 5 |
5’-CGCTGAGGAC AACACAGTCT-3’ |
5’-GCCACAGCCA GATTGAGAAC-3’ |
111 |
NA |
|
CD36 Exon 6 |
5’-TGTTCCAAGTCA GAACTTTGAGAG-3’ |
5’-CAGGGTACGGAA CCAAACTCA-3’ |
75 |
NA |
|
CD36 Exon5-6_6-7 |
5’-GGCAGCTGC ATCCCATATCT-3’ |
5’-CCATCTGCAGTA TTGTTGTAAGGA-3’ |
NA |
204 |
|
HPV16 L2 – CD36 intron 6 |
5’-TGCTGATGCAG GTGACTTTTAT-3’ |
5’-GCACCTTTCAC AATTTTTAAGGCCA-3’ |
NA |
212 |
|
HPV16 E2 - CD36 intron 5 |
5’-TACAGTGTCTAC TGGATTTATGTCT-3’ |
5’-AAACTTACCTCC GTACCAGTA-3’ |
NA |
160 |
|
HPV16 E6-E7 |
5’-GAACTGCAATG TTTCAGGACCCAC-3’ |
5’-ATTTCATCCTCC TCCTCTGAGCTG-3’ |
NA |
FL 578 E6*I 395 E6*II 278 |
Table 3: Primer sequences for PCR and/or RT-PCR of viral integration sites and affected genes.
2.7 Transcript Analysis
cDNA was prepared from RNA extracted from the FFPE blocks. cDNA was synthesized from 1mg of RNA using Superscript III and random hexamers. A No-RT control was prepared for each sample to ensure RNA purity. Primers were designed to amplify the native CD36 and LAMA3 transcripts proximal to and downstream of each viral integration site using NCBI Primer-BLAST (Table 3). Primers were also designed to amplify across the predicted fusion transcripts (Table 3). RT-PCR was performed using Platinum Taq DNA polymerase. Products of RT-PCR were separated and visualized with gel electrophoresis and were confirmed by Sanger Sequencing of the excised and purified bands.
3. Results
3.1 Case Report
A 60-year-old female nun in Cologne, Germany presented with bilateral p16-positive OPSCCs of the tonsils. The left tonsil was diagnosed as pT2N2bcM0, the right as pT2N0M0, both Grade 2. Combined radio-chemotherapy was recommended, but only radiotherapy was performed (59.5/50.4Gy) because the patient refused chemotherapy. The patient was free of disease at her last visit to the clinic two years post-diagnosis, after which she was lost to follow up but was reported to have died approximately one year later due to pneumonia.
3.2 HPV Testing
Staining for p16 was positive for both tumors, and HPV PCR-MassArray determined that HPV16 was the only HPV type present in the tumors (Figure 1).
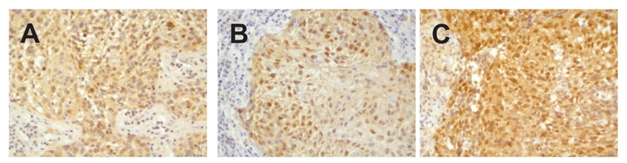
Figure 1: Immunohistochemical staining against p16INK4a. A) Left tonsillar tumor (block 1B). B) Right tonsillar tumor (block 2A) and C) Right tonsillar tumor (block 2D). V = 400x.
3.3 Integration analysis
DIPS-PCR revealed multiple integration sites in the tumor samples (Table 1). A total of six integration sites were identified; three viral integrations occurred in intergenic or genomic scaffold regions, and three viral integrations occurred in cellular genes (Figure 2A). The tumor from the left tonsil had integration of HPV16 L2 into intron 6 of the cellular gene CD36. Two blocks from the right tonsil were analyzed; the tumor cells from the right tonsil had integrations of HPV16 E5 into intron 5 of CD36, as well as an additional integration of HPV16 E5 into LAMA3, which likely caused a rearrangement of LAMA3 intron 1 and intron 68. It was curious to note that the segments of integrated HPV could all be aligned linearly starting from E1 to L2 (Figure 2B).
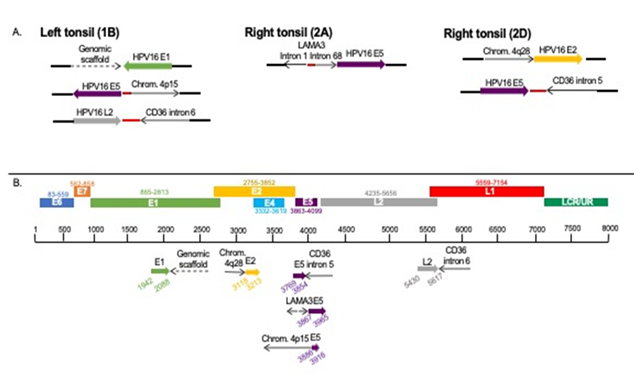
Figure 2: Viral integration sites identified by DIPS-PCR in each tumor block. The viral integration sites of HPV16 into the cellular genome. Red dots indicate unmappable sequence. A. Sites found within each sample. B. Sites from all samples aligned to HPV genome.
Direct PCR and Sanger sequencing confirmed the LAMA3 rearrangement and integration of HPV16 E5 occurred in the right tonsil, but it was not present in the left tonsil or the second block (Block 2D) from the right tonsil, suggesting this rearrangement did not persist in the other intratumoral clonal populations (Figure 3). Amplification of the native exons of LAMA3 DNA showed that there is an intact copy of LAMA3 exon 2 present in all 3 samples (Figure 4). However, only tumor 2A, which contained E5 integration into LAMA3, showed an intact copy LAMA3 exon 1.
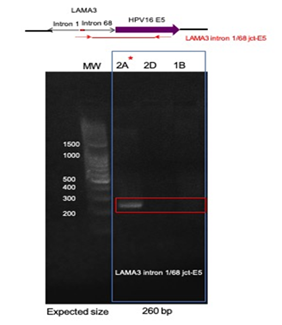
Figure 3: Confirmation of HPV16 integration into LAMA3. Primers designed within the junction of LAMA3 intron 1/intron 68 and HPV16 E5 were used to amplify the identified site in tumor DNA. PCR results, showing the predicted product in only tumor 2A, where the integration was originally identified (indicated by red star). 1B, left tonsil; 2A and 2D right tonsil blocks.
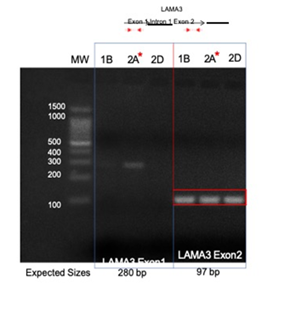
Figure 4: Amplification of native LAMA3 exons in tumor DNA. Primers within LAMA3 exon 1 and exon 2 were used to amplify tumor DNA. A product from exon 1 was present exclusively in tumor 2A (affected by LAMA3 integration), while all three tumors retained exon 2. 1B, left tonsil; 2A and 2D right tonsil blocks.
Direct PCR of the CD36 integration site found in the right tonsil (E5-CD36 intron 5) in the other blocks yielded no products; similarly, direct PCR of the site found in the left tonsil (L2-CD36 intron 6) in the right tonsil blocks yielded no products. Amplification of the native exons of CD36 DNA revealed exons 4, 5, and 6 were present in all blocks (Figure 5).
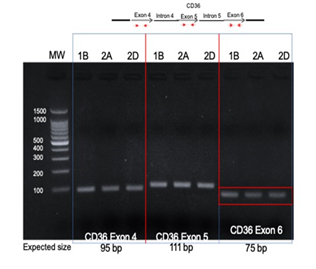
Figure 5: Amplification of native CD36 exons in tumor DNA. Primers within CD36 exon 4, 5 and 6 were designed and used for PCR with tumor DNA from each block. All tumors showed products at the expected sizes for each PCR reaction. 1B, left tonsil; 2A and 2D right tonsil blocks.
3.4 Transcript analysis
HPV16 E6-E7 transcripts were tested and were present in each sample (Figure 6A). Alternate E6-E7 transcripts were visible in each sample, but not full length E6-E7. The left tonsil tumor 1B, and one block from the right tonsil (2A) showed expression of E6*I-E7 and E6*II-E7, but block 2D only expressed E6*II-E7, consistent with evolution of the viral segments with tumor progression.
In order to understand whether viral integration into LAMA3 and CD36 disrupted expression of the genes, RT-PCR was performed. RT-PCR revealed that a transcript of the native LAMA3 exon downstream from the integration site (exon 2) was expressed in all 3 samples (Figure 6B). Normal CD36 transcripts spanning the two integration sites (the forward primer was designed to amplify the cDNA junction of exon 5-6 and the reverse primer to amplify from the exon 6-7 junction) were also expressed in all 3 samples (Figure 6A). These data suggest there is normal expression of at least one copy CD36 in each tumor or that the HPV L2 and HPV E5 were spliced out of the transcripts along with introns 5 and 6.
We attempted to amplify viral-host fusion transcripts that may have resulted from viral integration into LAMA3 or CD36. However, RT-PCR using primers designed to amplify the predicted fusion transcripts of LAMA3 and CD36 failed to yield any products. Every effort was made to limit the size of the amplicons, as FFPE RNA is usually highly fragmented making it difficult to amplify long products. Therefore, it is unclear whether these fusion transcripts are produced but unidentifiable due to fragmentation, or if they lack a viable promoter and are not expressed.
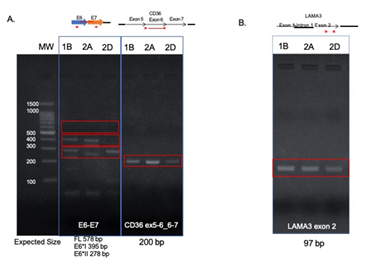
Figure 6: Transcript expression in FFPE tumor tissues. RT-PCR amplification of HPV16 E6-E7 (left) showed E6*I-E7 and E6*II-E7 in blocks 1B and 2A, but only E6*II-E7 in block 2D. RT-PCR amplification of the CD36 exons surrounding the integration sites (right) showed intact copies of CD36 in each sample. 1B, left tonsil; 2A and 2D right tonsil blocks.
4. Discussion
The incidence of HPV+OPSCC is rapidly rising in the Western world. Despite an overall survival advantage compared to HPV-negative cancers, there is still a significant proportion of patients who develop local or distant recurrences within 5 years and treatment de-escalation has failed [24, 25]. Therefore, there is a critical need to understand the cellular and molecular characteristics of HPV+OPSCCs with unfavorable outcome. Some studies have shown that viral integration of HPV into the genome is associated with worse prognosis [12, 15, 26] and these tumors have a different mutation signature, particularly of PIK3CA [27]. Viral integration into cellular genes may lead to disruption of gene expression and generation of viral-human fusion transcripts. Synchronous HPV+ bilateral tonsillar carcinomas are relatively rare with about 40 cases reported in the literature; however, studying their characteristics may contribute to our understanding of OPSCC [18, 21]. There is much controversy in the field whether all HPV+ tonsillar carcinoma patients should have their contralateral tonsil removed in order to prevent missing bilateral disease [28]. Others oppose this idea due to increased morbidity of removing both tonsils and lack of sufficient evidence this would benefit patients, especially given that most patients are given adjuvant radiation and monitored closely so disease in the contralateral tonsil could be caught [29]. The mechanism behind synchronous bilateral carcinoma development is debated; there is evidence for both clonal expansion of a single primary tumor as well as simultaneous development of independent carcinomas due to similar HPV exposure. Understanding how these tumors develop would help inform whether or not all patients with unilateral tonsillar carcinoma are at risk for development of bilateral disease.
Here we have presented a case of synchronous bilateral HPV+ tonsillar OPSCC from a 60-year-old nun. Although the patient claimed to never have sexual contact of any kind, the resulting tumors were HPV+. Viral integration analysis by DIPS-PCR of both tonsil tumors highlighted a number of HPV16 integration sites into both intergenic and genic regions of the cellular genome. There were viral integrations unique to each tonsil as well as within the right tonsil, for which two different areas of the tumor were analyzed. This included the integration of HPV16 E5 into the cellular gene LAMA3, which was only present in one block from the right tonsil, suggesting intratumoral heterogeneity.
The LAMA3 gene containing the viral integration was rearranged and inverted as a result of HPV16 E5 insertion as shown in Figure 2. The implications of the viral integration on LAMA3 function are unclear because no intact LAMA3 gene was present in blocks 1B and 2D. Limited analysis of the LAMA3 gene revealed that there is loss of LAMA3 exon 1 in the blocks without HPV E5-LAMA3 integration, but exon 2 is present in all three tumor areas. In trying to understand this observation, we postulated possibilities that might explain these results. If for example, we consider that the initial LAMA3 integration site was established via a “looping” integration mechanism which reversed the orientation of the LAMA3 gene and the integrated DNA, including LAMA3 exon 1, was subsequently excised during clonal expansion, this could explain why block 1B and 2D lost exon 1 DNA, but it is retained in block 2A. This suggests that the LAMA3 rearrangement included exon 1 and that this was an early event which was later excised as the tumor progressed in the right tonsil (block 2D) and the left tonsil (block 1B), but the only copy of LAMA3 carried in those tumor cells was the copy missing exon 1.
We also uncovered viral integration into the same cellular gene, CD36, in both tonsils. Despite the same cellular gene being involved, different but adjacent regions (E5-L2) of HPV were involved, and the integration sites were located in two different but sequential CD36 introns (intron 5 and intron 6). Although the locations of integration are slightly different, we expect it would be unlikely to discover viral integration into the same gene in unrelated samples; the majority of samples have unique viral integration sites due to the stochastic nature of viral integration [14, 30]. There is some evidence that viral integration occurs preferentially into genes [31], but it is unlikely that these integrations into CD36 are the result of separate viral integration events in two independent tumors. Furthermore, CD36 is not reported to be a hotspot of HPV integration, in contrast to other genes that have been reported in several cases [14, 32-34]. We believe that these two tumors arose from one primary tumor with viral integration into CD36 that underwent clonal expansion and was subsequently established in the other tonsil. However, the altered site of integration also suggests the DNA was edited with tumor evolution over time. It is unknown whether viral integration events are stable over time or if they are subject to changes due to either mobile element characteristics or genomic instability. It is also possible that the initial CD36 integration site was established via a “looping” integration mechanism and some of the integrated DNA was subsequently excised during clonal expansion, as has been described by others [35, 36].
LAMA3 encodes for the laminin subunit alpha-3, which is one of three members of the complex glycoprotein laminin 5. Laminins are components of the cellular basement membrane, and laminin 5 is reported to be involved in cell adhesion, migration, and the differentiation of keratinocytes [37]. Laminin 5 has also been shown to be overexpressed in invasive oral squamous cell carcinomas but not in premalignant lesions [38, 39]. CD36, or cluster of differentiation 36, encodes an integral membrane protein involved in fatty acid import and binds many ligands including collagen, lipoproteins, phospholipids and long-chain fatty acids. Studies done suggest CD36 may promote cell migration and proliferation in oral cancers and other solid tumors [40-42]. Our analysis suggests there is at least one intact copy of CD36, so the functional consequences are unclear. However, both CD36 and LAMA3 are involved in ECM-receptor interaction pathways and if their functions were somehow disrupted, this could provide a mechanism for tumor spread. Further investigation of CD36 and LAMA3 protein expression in these samples is warranted to understand whether these proteins are disrupted and whether that may have played a role in metastasis.
Our assessment of tumor clonality was limited to HPV genotype and HPV integration site, which other groups have also used to demonstrate tumor clonality [43, 44], but future work will include mutational profiling of cellular genes to strengthen our ability to assess the clonal nature of these samples. This work was somewhat limited by having access to only FFPE tissue, resulting in a lack of quality RNA for integrated viral transcript analysis. This limited our ability to detect gene expression changes of CD36 and LAMA3, as well as detection of the predicted viral-host fusion transcripts. It would have been valuable to evaluate the metastatic lymph node associated with the left pT2N2bcM0 tonsil, but DNA was not available for analysis from that metastatic lesion. Overall, our study supports clonal spread from one tonsil to another, and future work will be focused on validating these results in other bilateral tonsil pairs.
Acknowledgements
Studies carried out by the authors were supported by R01 CA194538 and P30 CA 046592. L.M. Pinatti was supported by the Cancer Biology Training Grant NIH-NCI Training Grant T32-CA009676. The authors would like to thank Nadine Jawad, Lila Peters and Christine Goudsmit for their assistance with sectioning and DNA isolation from tumor blocks, as well as Dr. John McHugh for his expertise on tumor pathology. We would also like to thank the University of Michigan Advanced Genomics Core for their assistance with Sanger sequencing.
Conflict of Interests
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
Contributions
L.M. Pinatti and H.M. Walline contributed to study conception and design, acquisition of data, analysis and interpretation of data, and drafting of manuscript; C.U. Huebbers and J.P. Klussman contributed to sample acquisition and clinical data, as well as study conception and design; T.E. Carey contributed to analysis and interpretation of data. All authors contributed to critical revision of the manuscript and gave approval for the final version.
References
- Centers for Disease Control, P. Centers for Disease Control and Prevention. Sexually Transmitted Diseases Treatment Guidelines: HPV-Associated Cancers and Precancers (2015).
- Chaturvedi AK, et al. Incidence trends for human papillomavirus-related and -unrelated oral squamous cell carcinomas in the United States. J Clin Oncol 26 (2008): 612-619.
- Brouwer AF, Eisenberg MC, Meza R. Age Effects and Temporal Trends in HPV-Related and HPV-Unrelated Oral Cancer in the United States: A Multistage Carcinogenesis Modeling Analysis. Plos One 11 (2016).
- Bray F, et al. Global cancer statistics. GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68 (2018): 394-424.
- Mehanna H, et al. Prevalence of human papillomavirus in oropharyngeal and nonoropharyngeal head and neck cancer--systematic review and meta-analysis of trends by time and region. Head Neck 35 (2013): 747-755.
- Chaturvedi AK, et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol 29 (2011): 4294-4301.
- Wittekindt C, et al. Increasing Incidence rates of Oropharyngeal Squamous Cell Carcinoma in Germany and Significance of Disease Burden Attributed to Human Papillomavirus. Cancer Prev Res (Phila) 12 (2019): 375-382.
- D'Souza G, McNeel TS, Fakhry C. Understanding personal risk of oropharyngeal cancer: risk-groups for oncogenic oral HPV infection and oropharyngeal cancer. Ann Oncol 28 (2017): 3065-3069.
- Mena M, et al. Might Oral Human Papillomavirus (HPV) Infection in Healthy Individuals Explain Differences in HPV-Attributable Fractions in Oropharyngeal Cancer? A Systematic Review and Meta-analysis. J Infect Dis 219 (2019): 1574-1585.
- Ortiz AP, et al. Periodontitis and oral human papillomavirus infection among Hispanic adults. Papillomavirus Res 5 (2018): 128-133.
- Pierce Campbell CM, et al. Long-term persistence of oral human papillomavirus type 16: the HPV Infection in Men (HIM) study. Cancer Prev Res (Phila) 8 (2015): 190-196.
- Walline HM, et al. Genomic Integration of High-Risk HPV Alters Gene Expression in Oropharyngeal Squamous Cell Carcinoma. Mol Cancer Res 14 (2016): 941-952.
- Akagi K, et al. Genome-wide analysis of HPV integration in human cancers reveals recurrent, focal genomic instability. Genome Res 24 (2014): 185-199.
- Parfenov M, et al. Characterization of HPV and host genome interactions in primary head and neck cancers. Proc Natl Acad Sci USA 111 (2014): 15544-15549.
- Huebbers CU, et al. Upregulation of AKR1C1 and AKR1C3 expression in OPSCC with integrated HPV16 and HPV-negative tumors is an indicator of poor prognosis. Int J Cancer 144 (2019): 2465-2477.
- Joseph AW, et al. Molecular etiology of second primary tumors in contralateral tonsils of human papillomavirus-associated index tonsillar carcinomas. Oral Oncol 49 (2013): 244-248.
- Roeser MM, et al. Synchronous bilateral tonsil squamous cell carcinoma. Laryngoscope 120 (2010): S181.
- Theodoraki MN, et al. Synchronous bilateral tonsil carcinoma: case presentation and review of the literature. Infect Agent Cancer 12 (2017): 38.
- Shimizu F, et al. Synchronous HPV-Related Cancer of Bilateral Tonsils Detected Using Transoral Endoscopic Examination with Narrow-Band Imaging. Case Rep Otolaryngol (2017): 9647010.
- Rasband-Lindquist A, Shnayder Y, O'Neil M. Synchronous bilateral tonsillar squamous cell carcinoma related to human papillomavirus: Two case reports and a brief review of the literature. Ear Nose Throat J 95 (2016): E30-E34.
- Rokkjaer MS, Klug TE. Prevalence of synchronous bilateral tonsil squamous cell carcinoma: A retrospective study. Clin Otolaryngol 43 (2018): 1-6.
- Walline HM, et al. High-risk human papillomavirus detection in oropharyngeal, nasopharyngeal, and oral cavity cancers: comparison of multiple methods. JAMA Otolaryngol Head Neck Surg 139 (2013): 1320-1327.
- Luft F, et al. Detection of integrated papillomavirus sequences by ligation-mediated PCR (DIPS-PCR) and molecular characterization in cervical cancer cells. Int J Cancer 92 (2001): 9-17.
- Mehanna H, et al. Radiotherapy plus cisplatin or cetuximab in low-risk human papillomavirus-positive oropharyngeal cancer (De-ESCALaTE HPV): an open-label randomised controlled phase 3 trial. Lancet 393 (2019): 51-60.
- Gillison ML, et al. Radiotherapy plus cetuximab or cisplatin in human papillomavirus-positive oropharyngeal cancer (NRG Oncology RTOG 1016): a randomised, multicentre, non-inferiority trial. Lancet 393 (2019): 40-50.
- Koneva LA, et al. HPV Integration in HNSCC Correlates with Survival Outcomes, Immune Response Signatures, and Candidate Drivers. Mol Cancer Res (2017).
- Leemans CR, Snijders PJF, Brakenhoff RH. The molecular landscape of head and neck cancer. Nat Rev Cancer 18 (2018): 269-282.
- Kim CM, St John MA. Should the Contralateral Tonsil Be Removed in Cases of HPV-Positive Squamous Cell Carcinoma of the Tonsil? Laryngoscope 129 (2019): 1257-1258.
- Cognetti D, et al. In Reference to Should the Contralateral Tonsil Be Removed in Cases of HPV-Positive Squamous Cell Carcinoma of the Tonsil? Laryngoscope 129 (2019): E194.
- Lace MJ, et al. Human papillomavirus type 16 (HPV-16) genomes integrated in head and neck cancers and in HPV-16-immortalized human keratinocyte clones express chimeric virus-cell mRNAs similar to those found in cervical cancers. J Virol 85 (2011): 1645-1654.
- Bodelon C, et al. Genomic characterization of viral integration sites in HPV-related cancers. Int J Cancer 139 (2016): 2001-2011.
- Hu Z, et al. Genome-wide profiling of HPV integration in cervical cancer identifies clustered genomic hot spots and a potential microhomology-mediated integration mechanism. Nat Genet 47 (2015): 158-163.
- Cancer Genome Atlas Research. Integrated genomic and molecular characterization of cervical cancer. Nature 543 (2017): 378-384.
- Li W, et al. Characteristic of HPV Integration in the Genome and Transcriptome of Cervical Cancer Tissues. Biomed Res Int (2018): 6242173.
- Groves I.J, Coleman N. Human papillomavirus genome integration in squamous carcinogenesis: what have next-generation sequencing studies taught us? J Pathol 245 (2018): 9-18.
- Holmes A, et al. Mechanistic signatures of HPV insertions in cervical carcinomas. NPJ Genom Med 1 (2016): 16004.
- Hamill KJ, Paller AS, Jones JC. Adhesion and migration, the diverse functions of the laminin alpha3 subunit. Dermatol Clin 28 (2010): 79-87.
- Rani V, McCullough M, Chandu A. Assessment of laminin-5 in oral dysplasia and squamous cell carcinoma. J Oral Maxillofac Surg 71 (2013): 1873-1879.
- Rahman F, et al. The expression of laminin-5 in severe dysplasia/carcinoma in situ and early invasive squamous cell carcinoma: an immunohistochemical study. Minerva Stomatol 62 (2013): 139-46.
- Nath A, et al. Elevated free fatty acid uptake via CD36 promotes epithelial-mesenchymal transition in hepatocellular carcinoma. Sci Rep 5 (2015): 14752.
- Hale JS, et al. Cancer stem cell-specific scavenger receptor CD36 drives glioblastoma progression. Stem Cells 32 (2014): 1746-1758.
- Sakurai K, et al. CD36 expression on oral squamous cell carcinoma cells correlates with enhanced proliferation and migratory activity. Oral Dis (2019).
- Arfi A, et al. HPV DNA integration site as proof of the origin of ovarian metastasis from endocervical adenocarcinoma: three case reports. BMC Cancer 19 (2019): 375.
- Lillsunde Larsson G, et al. HPV16 viral characteristics in primary, recurrent and metastatic vulvar carcinoma. Papillomavirus Res 6 (2018): 63-69.


 Impact Factor: * 5.3
Impact Factor: * 5.3 Acceptance Rate: 75.63%
Acceptance Rate: 75.63%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 
