Huaier Effects on Prevention and Inhibition of Spontaneous SARS-CoV-2 Virion Production by Repeated Pfizer-BioNTech mRNA Vaccination
Manami Tanaka1*, Tomoo Tanaka1, Fei Teng2*, Xiaolong Zhu2, Hong Lin2, Zhu Luo3, Sotaro Sadahiro4, Toshiyuki Suzuki5, Yuji Maeda6, Ding Wei7 and Zhengxin Lu8
1Bradeion Institute of Medical Sciences, Co., Ltd., Itado 433-1, Isehara, Kanagawa 259-1145, Japan
2BGI-Shenzhen, Building NO.7, BGI Park, No.21 Hongan 3rd Street, Yantian District, Shenzhen 518083, China
3BGI-Japan, KIMEC Center BLDG. 8F 1-5-2 Minatojima-minamimachi, Chuo-ku, Kobe 650-0047 Japan
4Department of Surgery, Kameda-Morinosato Hospital, 3-1-1 Morinosato, Atsugi, Kanagawa 243-0122, Japan
5Department of Surgery, Oiso Hospital attached to Tokai University School of Medicine, 21-1 Gakkyo, Oiso, Naka-District, Kanagawa 259-0198, Japan
6Department of Surgery, Kanagawa National Hospital, National Hospital Organization, 666-1 Ochiai, Hadano, Kanagawa 257-8585, Japan
7Japan Kampo NewMedicine, Co. Ltd., 2-8-10 Kayaba-Cho, Chuo-Ku, Tokyo 103−0025, Japan
8QiDong Gaitianli Medicines Co. Ltd., Jiangsu Province, China
*Corresponding Author: Fei Teng, BGI-Shenzhen, Building NO.7, BGI Park, No.21 Hongan 3rd Street, Yantian District, Shenzhen 518083, China.
Manami Tanaka, Bradeion Institute of Medical Sciences, Co., Ltd., Itado 344-1, Isehara, Kanagawa 259-1145, Japan.
Received: 06 January 2023; Accepted: 18 January 2023; Published: 24 January 2023
Article Information
Citation: Manami Tanaka, Tomoo Tanaka, Fei Teng, Xiaolong Zhu, Hong Lin, Zhu Luo, Sotaro Sadahiro, Toshiyuki Suzuki, Yuji Maeda, Ding Wei and Zhengxin Lu. Huaier Effects on Prevention and Inhibition of Spontaneous SARS-CoV-2 Virion Production by Repeated Pfizer-BioNTech mRNA Vaccination. Archives of Clinical and Medical Case Reports 7 (2023): 20-38.
View / Download Pdf Share at FacebookAbstract
Background: Although striking effects of vaccination strategy against pandemic COVID-19, a long-term influence by repeated partial virus mRNA injections are unknown. In addition, the molecular mechanism in accelerated ageing process still remains unclear. Through our clinical research, we observe and reported spontaneous SARS-CoV-2 virion and virion part production after Pfizer-BioNTech mRNA vaccination, observed from 3 weeks after the first injection. A significant destruction in ribosomal RNA structures, enhanced by repeated vaccinations, were also identified. The present study is aimed to define molecular mechanisms for SARSCoV- 2 virion production after injection of mRNA vaccination with a comparison to virion proliferation in non-vaccinated patient with severe COVID-19 pneumonia and fibrosis.
Methods: We direct clinical research to define molecular basis for significant anti-cancer efficacy of Huaier (Trametes robiniophila murr). In the present study, we used peripheral blood samples from the volunteer patient, suspected lung cancer by CT image analysis with age-matched normal controls with or without Huaier administration. Molecular characterization was performed analyzed by total RNA and non-coding small RNA sequencing on BGISEQ-500 Platform (about 7.0 GB analysis per sample). Thorough genetic events, Gene Ontology analysis, and functional target gene analysis were performed by using KEGGref) pathway classification (https://www.genome.jp/kegg/).
Results: Spontaneous SARS-CoV-2 virion production was identified after Pfizer-BioNTech mRNA vaccination until even 5 months after the 3rd vaccination. We compared this virion production in vivo with infected virus proliferation from severe COVID-19 pneumonia case without any vaccination by total RNA sequencing. No virion and virion particle were detected in normal c
Keywords
<p style="text-align:justify">Huaier (Trametes Robiniophila Murr); miRNA-medicated Transcription Control; mTOR/PI3K/AKT Signaling Pathway; Ribosomal RNA Structure; COVID-19 Signaling Pathway; SARS-CoV-2</p>
Article Details
Trial Registration
The Japanese Medical Association (ID: JMA-IIA00335, date of registration is on 1st February 2018).
1. Introduction
In the previous report [1], our clinical research demonstrated the evidence of SARS-CoV-2 virion and virion part in number even 3 months after the first vaccination by Gene Ontology (GO) classification and functional enrichment analysis of differentially expressed genes (DEGs). The long-term, over 2 years of influence of repeated mRNA vaccination against SARS-CoV-2 has not been clearly defined. It is noteworthy that, although Pfizer-BioNTech and Moderna vaccines contain mRNA that codes only for the SARS-CoV-2 spike protein [2], repeated vaccine shots promoted the production of many copies of SARS-CoV-2. Normal individuals usually can clear foreign genetic material within a few days, but SARS-CoV-2 production in vivo was detected from 3 weeks after the 1st vaccination, and persisted even 5 months after the 3rd vaccination. The influence of injected self-amplifying mRNAs, in addition to the subsequent spontaneous production, remains unclear. In addition, even with several vaccinations, there is still enough possibility of COVID-19 infection with novel mutated strains [3]. There is a certain limit on the preventive efficacy of repeated shots of anti-SARS-CoV-2 vaccination, and more importantly, the problems of serial shots were raised regarding the destructive effects on molecular mechanisms for protein synthesis by causing destructive ribosomal RNA structures [1, 4−7]. This raised many questions on the mechanisms, both in SARS-CoV-2 virions and in patient physiological systems, to induce the severity of the pathogenesis by SARS-CoV-2 infection. It is not the less important to emphasize that the problems of COVID-19 severe pneumonia exist not only in the disease itself but also in the severe and painful treatment strategies and consequent long-lasting complications [2, 3]. Currently we are launching into the postpandemic COVID-19 era and have found that more problems remain to be solved, such as more efficient preventive strategies and/or complications after invasive treatments. We have started genomic and genetic analysis of mRNA vaccination to our cancer patients from April 2020 [1, 8-15], and examined any alterations in genomic stability and in subsequent translation and transcription processes [12]. Our previous research results demonstrated that extensive genomic flexibility and capability were derived from opportunistic virus infections, and that those genomic potential is the basis to recover from the diseases [12]. Huaier (Trametes robiniophila murr), natural herb therapy on cancer patients (Chinese Administrative License No. Z-20000109) [8-15], contributes to making this genomic potential efficient for the rescue of disrupted physiological signaling networks. We found that Huaier could compensate for any damage from destructive ribosomal RNA structures after mRNA vaccination against SARS-CoV-2, dependent on the genomic potential of each individual, independent of the severity of cancer or basic health conditions [1]. The accelerated ageing process [16] in lipid metabolism has also been observed which affects micro-embolus production, causing increasing numbers of brain and cardiac infarctions in the Japanese population. In addition, it is unclear to distinguish the exact differences in the influence on physiological systems between by intrinsic/spontaneous viral particles by serial vaccinations, and by extrinsic/infected SARS-CoV-2 virus. The present study, together with the previous report [1], we succeeded in providing a clue and a solution for these problems, including a strategy for prevention and treatment without any complications. It is also indicated that the repair of destructive ribosomal RNA structures was s good indicator for the risk of spontaneous virion production as well as a good indicator of the recovery process. Molecular systems affected for these improvements were closely related to ageing process, via activation of mTOR/PI3K/AKT kinase-related signaling networks [12] as reported previously [1]. The novel KEGG analysis characterization (COVID-19 panel No. hsp05171) also demonstrated the influence of SARS-CoV-2 virions and virion part to lipid metabolism, too. A small dose of Huaier, 6 g per day, is enough to prevent SARS-CoV-2 infection as well as any unexpected damage in molecular systems in this normal control. Huaier effects contributed to control kinase function regulation via mTOR/PI3K/AKT signaling networks, cooperated with massive mi- and pi-RNA-medicated transcriptional control [1, 8-15]. The present study thus provides an efficient and non-hazardous strategy to cope with the up-coming postpandemic COVID-19 era.
2. Methods
2.1 Sample Collection and Medical Ethics Policy
Blood samples were taken from one patient, and two were healthy volunteers as controls. The present study was strictly conducted according to the guidelines of the Declaration of Helsinki and the principles of good clinical practice. Written informed consent was obtained from the patients. This clinical research was applied according to the Consolidated Standards of Clinical Research Trials guidelines and was registered with the Japanese Medical Association (ID: JMA-IIA00335, 1st February 2018). The project was strictly conducted with a review by the ethics committee consisting of experts on Medicine, Nursing, Laws, Pharmaceutics and Business Community (first committee held on 9th February, 2018) [8-12, 14-17]. Huaier (Trametes robiniophila murr) granules were prepared and provided by the original manufacturer (Chinese Administrative License No. Z-20000109), and maintained in the desired environment until use (within one year after production).
2.2 Quality Check of Total RNA Sample
An Agilent 2100 Bio analyzer (Agilent RNA 6000 Nano Kit) was used to perform the total RNA sample quality check (QC). The QC) item contains the RNA concentration, RIN value, 28S/18S and fragment length distribution.
2.3 Library and Sequencing
The obtained peripheral blood samples (nine samples from one cancer patient and two healthy controls) were designated for further sequencing. First, the poly-A containing mRNA molecules were purified using poly-T oligo-attached magnetic beads. Following purification, the mRNA was fragmented into small pieces using divalent cations under elevated temperature. After removal of rRNA, the RNA was converted into cDNA using reverse transcriptase and random primers. This step was followed by second strand cDNA synthesis using DNA Polymerase I and RNase H. These cDNA fragments then have the addition process by A-tailing and the cDNA was amplified. To generate libraries, the amplified products were separated into single strand DNA and cyclized. The RNA-seq libraries were subjected to sequencing with the pair-end option using the BGISEQ-500 system at Beijing Genomics Institute (BGI), China [15, 18]. We also performed total non-coding small RNA sequencing on the same platform. First, 18-30nt RNA segments were separated by PAGE. Ligation of corresponding adaptors was added to the RNA 5′ and 3′ ends, and the adapter-ligated small RNAs were subsequently transcribed into cDNA, and amplified product by several cycles. Then the RT primer was reverse-extended in next step to synthetize strand cDNA, and high-ping polymerase was used to amplify cDNA and enrich cDNA with both 3' and 5' adaptors. Purified DNA was used for cluster generation and sequencing analysis using the BGISEQ-500 platform. Total RNA was isolated from the serum samples and pooled together to construct cDNA libraries. These libraries were subsequently sequenced through Illumina HiSeq sequencing with paired-end reads of length 2*100bp according to the manufacturer’s instructions as previously described [15, 18].
2.3 Data Processing and RNA-seq Expression Analysis
The sequencing data were analyzed and filtered using software soapnuke software [19]. Clean reads were mapped to the human reference genome GRCh38 with software bowtie2 software. Then, the gene expression level of each sample was calculated using RSEM [20]. The expressed genes were further analyzed between the groups and were detected by DESeq2 [21] software. Significant differentially expressed genes (DEGs) were defined as those with a fold-change larger than 2 and a p-value smaller than 0.05.
2.4 Data Processing and Expression Analysis of miRNAs
Low-quality sequencing data were removed first, and the reads were aligned to the miRbase database with bowtie2 [22] software, and the expression level of miRNAs was calculated and standardized by TPM. Differentially expressed miRNAs between two samples were screened strictly based on Poisson distribution. Then, we made multiple hypothesis test correction for the p value of the difference test, and to judge the significance of gene expression difference, FDR≤ 0.001, and the absolute value of Log2Ratio≥1 was set as the default threshold. MiRanda [23] and TargetScan [24] software were used to obtain the target gene of differentially expressed miRNAs and to extract the intersection or union of the target genes as the final prediction result. The metabolic and signal transduction pathways and their biological functions were determined. Pathway enrichment and GO significance enrichment analyses were performed on the target genes of differentially expressed miRNAs or DEGs by using the R package ‘phyper’. For each P-value, we corrected for multiple comparisons by controlling the False Discovery Ratio. The terms for which the FDR was not greater than 0.01 were defined as statistically significantly enriched.
3. Results
3.1 Sampling Time Course and COVID-19 Patient Characterization
Over hundreds of analysed samples in our clinical research [1, 8-15], we hired 3 types of candidate volunteers for the present study. Samples from two healthy Japanese males were used as normal controls with (6 g per day since November 2022) or without Huaier administration. The characterization of total RNA sequencing data of both have been reported and used in a series of our reports [1, 8-15]. The other was a severe COVID-19 pneumonia patient treated with Huaier at 30 g per day for 3 months. Sampling timing was as follows; 1) before 1st vaccination, 2) 3 weeks after 1st vaccination (just before 2nd vaccination), 3) 3 weeks after 2nd vaccination (just before 3rd vaccination), 4) 3 months after 2nd vaccination, 5) 6 months after 2nd vaccination, 6) 9 months after 2nd vaccination (just before 3rd vaccination), 7) 3 weeks after 3rd vaccination, 8) 5 months after 3rd vaccination (just before 4th vaccination), and 9) 3 weeks after 4th vaccination (data was still under analysis). Currently, the 5th vaccination is not planned in these two individuals. The Pfizer-BioNTech mRNA vaccine was used throughout the present study. In the present study, we used total RNA sequencing method including small nuclear RNAs, with quantitative and qualitative functional analysis. The data presented were from the timing of 5 months, 9 months after the 2nd vaccination, since the former data until the 3rd vaccination have been presented in a previous report1. These samples were used to show the influence of repeated vaccination and subsequent intrinsic/spontaneous virion production in vivo. To compare the molecular information from vaccinated individuals with extrinsic/infected virus proliferation, samples from one patient (68-years-old male) with severe pneumonia were used. The patient had no vaccine shots at all. To introduce the case details, the CT image analysis (Figure 1) and blood test results are summarized in Table 1. The onset time of infection was suspected in the middle of January by the history of close contact with the severely infected patient with the SARS-CoV-2 Omicron strain. Symptoms were heavy cough, sputum, and chest pain at night, which prevented him from sleeping and body weight loss by 10 kg within 4 weeks, with blood oxygenation levels of 86-87 % (detected by right finger) and 90% at most (left finger). No fever was detected throughout the observation period. Suspected as lung cancer by CT image analysis, Huaier therapy was applied at 30 g per day. Within a couple of weeks, drastic improvement was observed, and almost all the symptoms were eliminated. No fever was detected throughout the treatment period of 3 months. The improvement was confirmed by CT image analysis (Figure 1), which eradicated the multiple infiltrate shadows in both lungs. A few linear and vitreous shadows remained even at the end of June. As shown in Table 1, a significant efficacy to indicate the process of recovery was emphasized by the biomarker KL-6 (Mucin short variant S1) [25-27], designated to be a laboratory parameter for adenocarcinoma and pulmonary fibrosis (interstitial pneumonia).
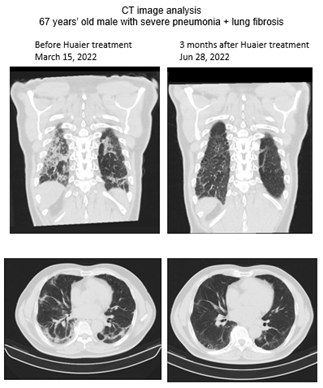
Figure 1: Chest CT image analysis before (March 15) and after 3 months of Huaier treatment on the patient (June 28).
|
Date of blood sampling |
Mar. 15, 2022 |
May 30, 2022 |
|||
|
measured titer |
measured titer |
Standard values |
|||
|
High or Low |
High or Low |
||||
|
SAARS-CoV-2 IgG |
not tested |
6.83 |
<1.40 |
||
|
KL-6 |
1,222 |
H |
610 |
H |
<500 |
|
CRP |
0.82 |
H |
0.97 |
H |
<0.30 |
|
WBC |
10,090 |
H |
10,790 |
H |
3,500~9,700 |
|
RBC |
476 |
508 |
436~577 |
||
|
Hb |
13.4 |
14.3 |
13.6~18.3 |
||
|
Ht |
43.1 |
46.7 |
40.4~51.9 |
||
|
MCV |
91 |
92 |
83~101 |
||
|
MCH |
28.2 |
28.1 |
L |
28.2~34.7 |
|
|
MCHC |
31.1 |
L |
30.6 |
L |
31.8~36.4 |
|
Plt |
39.9 |
H |
38.3 |
H |
14.0~37.9 |
|
Baso |
0.6 |
0.5 |
0.0~2.0 |
||
|
Eosino |
4.8 |
3.3 |
0.0~7.0 |
||
|
Neutro |
59.9 |
63.7 |
42.0~74.0 |
||
|
Lympho |
28.3 |
27.6 |
18.0~50.0 |
||
|
Total protein |
7.3 |
7.6 |
6.5~8.2 |
||
|
Total Bil |
0.5 |
0.3 |
0.3~1.2 |
||
|
ALP |
71 |
38~113 |
|||
|
LDH |
181 |
120~245 |
|||
|
AST (GOT) |
20 |
16 |
10~40 |
||
|
ALT(GPT) |
18 |
13 |
5~45 |
||
|
g-GTP |
26 |
20 |
<79 |
||
|
CPK |
66 |
87 |
50~230 |
||
|
Amylase |
79 |
73 |
39~134 |
||
|
TG |
231 |
H |
222 |
H |
50~149 |
|
HDL-C |
35 |
L |
33 |
L |
40~80 |
|
LDL-C |
91 |
85 |
70~139 |
||
|
UN |
15 |
20 |
8.0~20.0 |
||
|
Creatinine |
0.77 |
0.83 |
0.65~1.09 |
||
|
UA |
8.2 |
7.1 |
H |
3.6~7.0 |
|
|
Na |
142 |
143 |
135~145 |
||
|
K |
4.8 |
4.8 |
3.5~5.0 |
||
|
Cl |
105 |
106 |
98~108 |
||
|
Blood Sugar |
116 |
H |
98 |
70~109 |
|
Table 1: The blood test results comparison between before (March 15) and after 3 months of Huaier treatment on the patient (June 28).
3.2 Destructive Ribosomal RNA Structures Identified in the Patient and Normal Control after the 3rd Vaccinations (without Huaier Administration)
Here, we demonstrate the obtained results by the process of laboratory analysis, i.e., 1) starting from total RNA extraction from the blood samples, 2) RNA quality check, 3) cDNA library construction and total sequencing (including non-coding small RNA sequencing on the same platform), 4) data processing and expression analysis of RNA and small nuclear RNAs, and 5) functional analysis of target genes and differentially expressed genes (DEGs) and differentially expressed small nuclear RNAs (DSGs). Extracted RNAs were designated for quantifying RNA concentration, 28S/18S & 23S/16S ribosome structures, and RNA integrity number (RIN) or RNA quality number (RQN) values for further library construction. Figure 2, left column in each panel, demonstrates the resulting ribosomal RNA structures observed throughout the research period in the present study. As the same as samples from severely advanced cancer patients1, 8, the samples evaluated as risky or unqualified for further processing are indicated by an asterisk (*) in Figure 2, panels A and C). Surprisingly, a remarkable similarity in the destructive pattern was observed between the first sample of the patient (before Huaier administration; Figure 2, panel C) and that in the normal control-1-3, 5 months after the 3rd vaccination (Figure 2, panels A and C; indicated by *). The destructive pattern identified in the patient was improved and recovered significantly by the time course of Huaier administration, and became within normal limits after 3 months of treatment (Figure 2, panel C, left column). Normal control No.2 (Figure 2, panel B) did not show any destructive patterns. He had continuous Huaier administration at a low dose of 6 g per day from November 2020. Without Huaier administration (normal control No. 1; Figure 2, panel A), progressive destruction was obviously detected, such as the 5S peak on the high side, which affects the quantitative inaccuracy and is conducive to the inaccuracy of the loading amount and the poor data quality. The RIN or RQN values were slightly under standard, too, with the same poor quality of 28S/18S or 23S/16S values evaluated also as under standards.
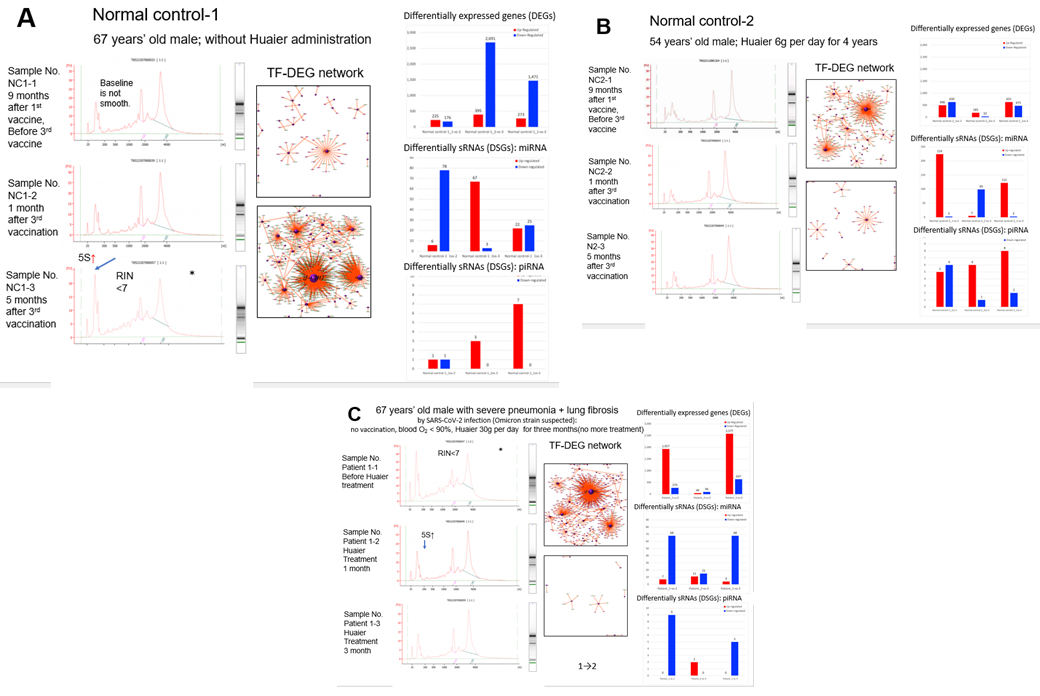
Figure 2: Ribosomal structures by HPLC analysis (left column), with TF-DEG network (middle column). The time of sampling, and Pfizer-BioNTech vaccinations (panel A. normal control 1; panel B. normal control 2; panel C for patient data) are indicated on the left side of each panel. The red and green dots in the TF-DEG network represent the up-regulated and down-regulated DEGs, respectively. The purple ball represents a transcription factor, and the greater the node is, the more DEGs the transcription factor regulates. In the right column of each panel, a comparison of the numbers of differentially expressed genes (DEGs) was placed according to the time course of Pfizer-BioNTech vaccination; red bars represent up-regulated molecules, and blue bars represent down-regulated molecules.
3.3 Extensive Genomic Plasticity and Flexibility demonstrated by the Repeated Pfizer-BioNTech mRNA Vaccination and in the First Month of Huaier Treatment
A summary of the sequence results and RNA editing events is shown in Supplementary Extended Table 1. As noted in a series of publications by our clinical research [1, 8-15], extensive genomic plasticity and flexibility were observed. The details of the genetic and genomic characterization of those two normal controls were also emphasized and highlighted in the previous report [1]. However, the patient did not demonstrate significant differences among the sequence events compared with the observation in cancer patients already reported [8-15]. For example, we identified 135,700 SNP variant types in the SARS-CoV-2-infected patient in the sample analysis in mean number ranging from 128,033 to 144,654 in the patient, whereas 22,688 in total among normal healthy individuals [18] without Huaier or mRNA vaccination (Supplementary Extended Table 1). The significant increase in total SNP numbers was consistent with the precious reports. Virus infection did not seem to affect the significant increase in SNP numbers. For the SNP variations, A*G > C*T(I) transitions were the most common mutations (301,899, 74.9%), followed by, C*G>G*T(I) (57,376, 14.2%) transversions, and A*C>A*T (I) (43,563, 10.8%) transversions, which is consistent with the data obtained among the cancer patients provided in the previous reports, and normal healthy controls with mRNA vaccine shots. As a result of these analyses, gene expression level analysis enabled us to compare the levels of differentially expressed genes (DEGs) among samples (detailed processing data available in the BGI database; see Methods section). We analysed the quantitative and qualitative alterations of all the transcriptomes, DEGs and DSGs. The results demonstrated these DEG relationships and the encoding transcription factors (TF) network analysis was performed [1, 8-15], and the results are shown in the middle column of Figure 2 panels. In the right column, quantitative changes in up/downregulation of DEGs and DSGs (mi- and pi-RNAs) were added in each panel (numbers of altered genes are written on the top of each bar). No siRNA alterations were detected in the present study. Drastic DEG changes were observed together with a super-nova demonstration in the TF-DEG network, which were equivalent to the alteration quantity level detected in the cancer patients [11-13] (Figure 2, panel C). As noted before [13], viral infection of the patient evokes at most changes in genetic alterations. The numbers of upregulated DEGs were 2,577 in total at 3 months, approximately 10% of the total transcriptome numbers. In addition, more inhibitory effects, down-regulation of transcriptomes were overwhelmed than the numbers of up-regulated ones (Figure 2, panel C). Alterations in DEGs were the results of almost all up-regulation, whereas the alterations in DSGs were down-regulated. The results in DEGs and DSGs were controversial, together with the similar results obtained by analyzing cancer patient samples. Such contradictory changes between DEGs and DSGs should be considered sRNA-mediated transcriptional control to compensate for the physiological influence by virion and virion particles, compared with the various types of Huaier therapy on cancer patients and normal controls in the previous reports [11-13]. The results of the changes by TF-DEG network (Figure 2, panels B and C, middle column) clearly demonstrated the on-demand style of Huaier compensation of physiological systems. The drastic DEG alterations were evoked using transcriptional factors as network centers, and ceased after enough activity to rescue the impaired functions. It was most typically demonstrated at the first month of Huaier treatment of the patient (up-regulation) (Figure 2, panel C), after the 3rd vaccination of normal control 1 (down-regulation) (Figure 2, panel A), and at every period of normal control 2 (almost no alterations required) (Figure 2, panel B). In these results, again, the up- and down-regulation style was controversial between DEGs and DSGs. No specific transcription factors or groups of factors were identified in the present study, or throughout our clinical research. However, massive mi-RNA up-regulation was identified in normal control 2, even though quantitative DEG alterations were minimal (Figure 2, panel B). Here we have another question; if the significantly low degree of the TF-DEG network changes in Figure 2, how can the possible damage from destructive ribosomal RNA structures, which might affect translational and transcriptional processes, be restored and maintained? Of course, we remember the genetic potential observed in the drastic increase in SNP/INDEL variants and total abundance of isoforms described above. We should identify how and why silenced TF-DEG network could recover the damages. The role of DSGs and pi- and miRNA qualitative analysis are shown in Table 2.
Table 2: A list of miRNA and piRNA regarding to the transcriptional control on DEGs identified in the normal controls 1-2 and the COVID-19 patient by the time course of Huaier administration. The comparisons of DSG level are written by Log 2 ratio. The real numbers of up/down-regulated mi- and piRNAs are shown in all panels in Figure 2 right column. The comparison is made between the data analysed in sampling 1 and 2 or 3. For normal controls, 1; before the 3rd vaccine, 2; 1 month after the 3rd vaccination, 3; 5 months after the 3rd vaccination. For the data from the patient, 1; before Huaier administration, 2; 1 month after Huaier administration, 3; 3 months after Huaier administration. Red bar represents upregulated sRNAs and blue bar represents downregulated sRNAs. Upper panel: miRNA and lower panel: piRNA.
3.4 KEGG Pathway Analysis on COVID-19 Signaling Cascade (hsa05171) regulated by the Integrated Kinase Signaling Network (mTOR/PI3K/AKT Signaling Cascades)
Here we proceed into the functional analysis of targeted genes. With quantitative analysis of DEGs (Figure 2, right column of each panel), we performed KEGG pathway classification (https://www.genome.jp/kegg/) to define the detailed mechanism on SARS-CoV-2 infection and regulating systems [28]. Recently, another KEGG panel of COVID-19 signaling cascades has become available (hsa05171). Figure 3 panel A summarizes hsa05171 analysis of the patient with a comparison to two normal controls with or without Huaier administration. The results revealed a strikingly minimum change in DEGs, whereas gross alterations are shown in Figure 2 (top of right column in each panel). Notable changes were identified in the expression of PI3K and structures of the 80S ribosome construct. The next step was to obtain detailed information about phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K) and related integrated kinase signaling networks. We demonstrated the mTOR/PI3K/AKT signaling cascade (KEGG pathway map04150) [28-34] in Figure 3 panel B. Here, we clearly show the molecular basis of recovery or rescue of function in the patient. In contrast, the failure of functional compensation was typically demonstrated in normal control 1; the transcripts were significantly down-regulated and functional genes were not activated, which also provided a molecular basis fora the observation of the progressive destructive changes in ribosomal RNA structures. As expected, a minimum essential increase in kinase function was observed in normal control 2, even though enough genetic alterations in transcriptional factors were detected. The mTOR/PI3K/AKT kinase-regulated signaling pathway [16-17, 34-37], reported to be responsible for the regulation of ribosomal RNA synthesis, showed a significant relationship to the results demonstrated by using the KEGG panel hsa05171 (Figure 2 panel A). With or without Huaier administration, restoration and activation of PI3K and AKT kinase function contribute to the maintenance of normal cellular signaling networks. Massive inhibition and down-regulation of related factors in PI3K and AKT kinase observed in normal control 1 has been reported to result in poor prognosis in cancer patients. As shown in Figure 3, the activation of neural signal transfer should be closely related and required for the maintenance of cellular mechanisms. Thus, the efficacy of Huaier administration seemed to be closely related to the rescue and repair of damaged functions after SARS-CoV-2 infection. The defects in lipid metabolism and neuro-degenerative factors also caused the disorders appearing as a long-term influence after mRNA vaccination (highlighted by red or blue box).
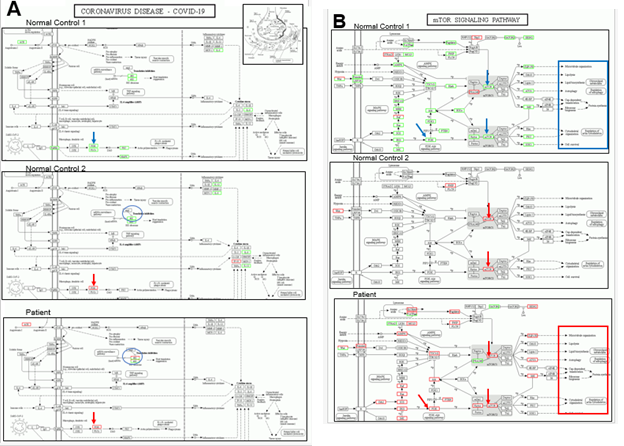
Figure 3: KEGG pathway analysis on COVID-19 panel hsp05171 (panel A) and transcriptional misregulation and the P13K/AKT signaling pathway panel B). Huaier administration was applied in the patient and normal control No.2. (5 g/day). Red bars represent up-regulated molecules, and blue bars down-regulated molecules. The figure was chosen at the time of the highest transcription factor-DEG network flexibility in each patient among serial analyses over a long time course of treatment.
3.5 GO Functional Analysis on Cellular Mechanisms
Based on the DEG and KEGG pathway annotation [34], Figure 4 shows the pathway and process enrichment analysis, both quantitatively and qualitatively to clarify the influence of intrinsic and extrinsic virion and virion particles. Various pathway classifications and functional information contributing to the expression changes were demonstrated. In the middle column in Figure 4 panel A (cellular component section), virion and virion particle numbers were noted. As clearly shown, virion production in vivo was strongly similar to that in the first panel of patients (Figure 4, panel B), indicating proliferation of the extrinsic/infected virus. Interestingly, no virion were detected in normal control 2 with daily Huaier administration. The results were consistent with the destructive ribosomal RNA patterns. It seems that the numbers of virions were proportional to the extent of destruction of ribosomal RNAs, and decreased by the process of recovery. Again, we could detect virion particles produced by mRNA vaccination against SARS-CoV-2 [1], which is designated to produce only part of the spike of the virus. Huaier administration activates the cellular function repair systems in total, but as shown in Supplementary Extended Table 2, it is unlikely to contribute to the significant influence on the molecules responsible for iPS/ES production to enhance tissue regeneration and repair systems, such as those identified in the patient lungs.
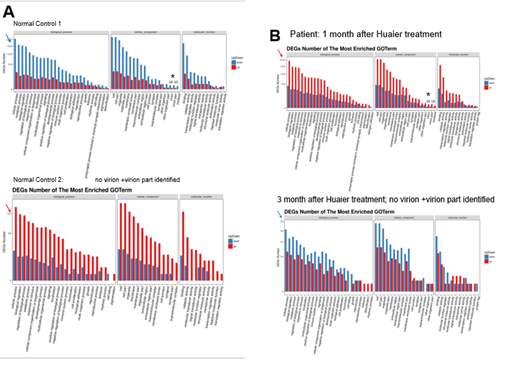
Figure 4: The GO enrichment pathway analysis of all the DEGs at 5 months after the 3rd vaccination (normal controls 1 and 2 in panel A), and by the time course of Huaier treatment on the patient (panel B). The X axis is the terms of pathway, and the Y axis is the number of increased or decreased genes. The numbers of virion and virion part was indicated by * with numbers in each lane. Red bars represent up-regulated molecules, and blue bars represent down-regulated molecules. Arrows indicate the quantitative changes in up-regulated (in red color) and down-regulated (in blue color) DEGs.
3.6 Significant Quantitative and Qualitative Alterations in DSGs (mi- and pi-RNA)
In the present study, small non-coding RNAs (sncRNAs) were simultaneously sequenced using BGISEQ-500 technology [12, 35]. The statistics of detected small non-coding RNA (DEGs) and miRNAs are shown in the upper panel of Table 2, and piRNAs are shown in the lower panel throughout the research period. No altered siRNAs were identified in the present study. The detailed numbers of each sample are already shown in Figure 2 panels. The significant and drastic increase in down-regulated DESs (blue-bars) matched the significant alterations in transcriptional factors (total 2,216), as noted before. However, many miRNAs listed in Table 2 had no significant and meaningful functional relations to the recovery of COVID-19 infection. Each individual showed specific patterns of DSG alteration style, and it is noteworthy to detect that the drastic up-regulations of DSGs in normal control 2 resulted in minimum alterations in DEGs [36]. The list of DEG alterations is listed in Table 2. The piRNAs with the lowest log2 ratio were identified in the patient within 1 month of Huaier administration, i.e., in order of decreasing number: hsa_piR_020582, 020496, 015249, 016742, 016658, 000805, 016659, 016677, 004150 (in the order of decreasing log2 ratio. After 3 months of treatment, 016735, 020496, 016658, 016659, and 020582 (020496, 016658, and 016659 were found throughout the treatment period). However, those sequences were still functionally unknown, with no possible relationship to any of the encoded TFs. We could only note the controversial relationship between up-regulation and down-regulation in DEGs and DSGs (especially in miRNAs).
4. Discussion
Thus, adjuvant therapy with Huaier, kinase function regulator [1], in rescuing the defects caused by destructive ribosomal RNA structures by massive genomic flexibility and capability, which might be consistent with SARS-CoV-2 virion and virion part production in vivo. As represented by the excessive flexibility observed in SNP and splice-type variants, we have provided much evidences that virus infection, including opportunistic infections, might cause at most genomic changes [13]. These are not simple processes, and genome-wide mobility seems to matter the consequent genetic events. The molecular ability of the functional compensation was well demonstrated as TF-DEG network (Figure 2, middle column), which consequently resulted in the recovery from the accelerated ageing process in lipid metabolism [16, 38], hair growth and tissue regeneration [17], inter/intra neural signal transfer [17], and endocrine secretion systems [8-10]. Considering the inhibitory influence on DEG management even with 3rd vaccine shots in normal control 2, minimum essential energy consumption was enough for this compensation. The present study revealed spontaneous virion production after repeated mRNA vaccination in normal control 1 without Huaier administration (Figure 4 panel A). Although Huaier does not prevent or influence virus infection itself, the efficacy of Huaier observed in normal control 2 can be speculated to control the latent damages of viral particle existence by down-regulation of production, and that with a small dose of 6 g per day. Ribosomal RNA is also the target of numerous clinically relevant antibiotics [37], however, the influence of mRNA vaccination itself has not yet been reported. According to our sequential analysis, we should observe a long-term influence of low quality ribosomal RNAs after repeated mRNA vaccinations by year. The present study, however, provided evidence up-regulation of kinase AKT and its mediating activation of the mTOR/PI3K/AKT signaling pathway rescues this possible damage [39-42]. The PI3K/AKT/mTOR pathway is an intracellular signaling pathway important in regulating the cell cycle. Therefore, it is directly related to cellular quiescence, proliferation, carcinogenesis, and longevity. TPI3K activation phosphorylates and activates the kinase AKT [43]. AKT can have a number of downstream effects such as activating CREB [30], inhibiting p27 and p21 [44], for further transcription factor mediated functional control of the cells, and activating the mTOR signaling pathway [44]. There are many known factors that enhance the PI3K/AKT pathway including EGF [45], sonic hedgehog [30], IGF-1 [30], insulin [44], and CaM [45]. The pathway is antagonized by various factors including PTEN [46], GSK3B [30], and HB9 [32]. In the present study, we focused on the significance of KL-6 as a biomarker in COVID-19 pneumonia and fibrosis. KL-6, mucin short variant S1, also called polymorphic epithelial mucin (PEM) or epithelial membrane antigen (EMA), is a mucin encoded by the MUC1 gene in humans [38]. Mucins line the apical surface of epithelial cells in the lungs, stomach, intestines, eyes and several other organs [25], which serves a protective function by binding to pathogens [26] and functions in a cell signaling capacity [27]. Overexpression and other modifications in the protein have also been associated with carcinogenesis. The Huaier effects on lung tissue repair observed in the present study also analysed DEG alterations in the 6 genes that will be responsible for promoting growth and proliferation over differentiation of stem cells. However, we could not observe a significant relationship (Supplementary Extended Table 2). It can be speculated that 1) Huaier could compensate for any damage from destructive ribosomal RNA structures, but at the same time, 2) the extent of compensation depended on the genomic potential of each individual independent of the severity of cancer or basic health conditions, and/or massive up-regulation in all these molecules responsible for iPS/ES cell productions were required. Pfizer-BioNTech and Moderna vaccines contain mRNA that codes for the SARS-CoV-2 spike protein. The present study revealed that, when the vaccines are injected, they deliver the mRNA to cells, which makes copies of not only expected spike, but also promotes spontaneous virion production. Huaier administration induced significant down-regulation of those virions and derived particles, which was not dependent on the dose of Huaier. It has been reported that a self-amplifying vaccine will be introduced in additional to the vaccine strategy against SARS-CoV-2, and others in development include enzymes from alphaviruses to repeatedly copy the genetic strand inside a cell and stay in the body for more than twice as long. The influence of the self-amplifying mRNA COVID-19 [47] vaccine, together with the spontaneous copies of virion production remains unclear, and we need a scientific answer for a novel vaccine strategy to overcome this long-term influence of (repeated) vaccination. Although we can speculate the possible relationship between the presence of spontaneous SARS-CoV-2 virion production and destructive ribosomal RNA structures, the role of the self-producing SARS-CoV-2 virion particles, and its destructive effect remain unknown. The results obtained here also showed that normal control 1 decided to have Huaier administration (20 g per day) 3weeks after the 4th vaccination. We are expecting the recovery and modification of the damaged ribosomal RNA structure by observing massive hair growth within one month of Huaier treatment. There were no specific miRNAs and piRNAs related to destructive ribosomal RNA structure production, whereas a couple of reports mentioned that a certain group of DSGs were related. We identified massive down-regulated DSGs (Table 2) that have been reported previously to be typical observations even in cancer patients as well as normal healthy controls with Huaier administration. It seems that these effects were not dependent on the dose, unlike the dose-dependent efficacy of Huaier. Although the functions of piRNAs are still poorly understood, piRNAs as novel attack tools against COVID-19 are being reported [48]. In contrast, the alterations in miRNAs were closely related to transcriptional factor chances as shown in Figure 2 panels and Table 2, which might contribute to decreasing the damage and dysfunction caused by destructive ribosomal RNAs.
5. Conclusions
Consequently, the present study again emphasizes the significance of Huaier adjuvant therapy on any strategy against COVID-19, with or without repeated vaccination, as a non-hazardous kinase regulator. These results are encouraging to any human society toward the up-coming postpandemic COVID-19 era. The data after the 4th vaccinations, and the rescue of disrupted ribosomal RNA structure in normal control 1 by Huaier treatment will follow.
Data Availability
The complete sequencing data of the cases have been deposited into the NCBI database, and the clinical outcomes of these cases are not publicly available for data privacy but are available from Dr. Fei Teng: Sky.teng@bgi.com) and Dr. Manami Tanaka (e-mail: tubu0125@gmail.com, manami-tanaka@bradeion.com upon request for research collaboration. The timeframe for response to data access requests is 30 days. There are no restrictions on the reuse of data. In addition, the raw data of the longitudinal cohort and healthy individuals analyzed in this study were available at GEO with identifiers of NCBI GEO (GSE157086).
Acknowledgements
The authors wish to thank the patient and normal healthy volunteers who kindly collaborated with the present study.
Funding
The present study was a grant-in-aid from QiDong Gaitianli Medicines Co., Ltd. and Japan Kampo NewMedicine, Co., Ltd. The request for Huaier provision from all over the world should be addressed to: Dr. Zhengxin Lu, QiDong Gaitianli Medicines Co. Ltd., Jiangsu Province, China. E-mail: lzx2777@163.com.; lyh307@sina.com.
Contributions
T., and T. T. designed the study from the clinical observation of cancer patients with Huaier treatment (as a complementary therapy), managed the sampling and clinical assessment of the patient volunteers, statistically analyzed the data, and drafted the manuscript. F. T., X. Z., and H. L. Z. L., managed total RNA and small nuclear RNA sequencing and conducted systematic analysis of the data. S. S., T. S., and Y. M., contributed to the clinical diagnosis and treatment of the patients, together with the assessment of QOL and the effects of Huaier administration, Z. L., D. W., contributed to the provision of Huaier granules and clinical evaluation of the data, especially focused on immunological evaluation.
Ethics Approval and Consent to Participate
Blood samples were taken from one patient, and two were healthy volunteers as controls. The present study was strictly conducted according to the guidelines of the Declaration of Helsinki and the principles of good clinical practice. Written informed consent was obtained from the patients. This clinical research was applied according to the Consolidated Standards of Clinical Research Trials guidelines and was registered with the Japanese Medical Association (ID: JMA-IIA00335, 1st February 2018). The project was strictly conducted with a review by the ethics committee consisting of experts on Medicine, Nursing, Laws, Pharmaceutics and Business Community (first committee held on 9th February, 2018) [8-12, 14-15, 30-31]. Huaier (Trametes robiniophila murr) granules were prepared and provided by the original manufacturer (Chinese Administrative License No. Z-20000109), and maintained in the desired environment until use (within one year after production).
Consent for Publication
Not applicable.
Competing Interests
The authors declare no competing interests.
References
- Tanaka M, Tanaka T, Zhu X, et al. Huaier effects on functional compensation with destructive ribosomal RNA structure after anti-SARS-CoV-2 mRNA vaccination. Arch Clin Biomed Res 6 (2022): 553-574.
- Chakraborty S, Mallajosyula V, Tato CM, et al. SARS-CoV-2 vaccines in advanced clinical trials: Where do we stand? Adv Drug Deliv Rev 72 (2021): 314–338.
- WHO, 2021. Coronavirus Situation Report (2021).
- Chan JC, Hannan KM, Riddell K, et al. AKT promotes rRNA synthesis and cooperates with c-MYC to stimulate ribosome biogenesis in cancer". Sci Signal 4 (2011): ra56.
- Hoppe S, Bierhoff H, Cado I, et al. AMP-activated protein kinase adapts rRNA synthesis to cellular energy supply. Proc Natl Acad Sci USA 106 (2009): 17781-17786.
- Liang XH, Liu Q, Fournier MJ. Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing. RNA 15 (2009): 1716-1728.
- Brandman O, Hegde RS. Ribosome-associated protein quality control. Nat. Struct. Mol. Biol 23 (2016): 7-15.
- Tanaka M, Tanaka T, Teng F, et al. Huaier compensates impaired signal transfer inter/intra neurons in central and peripheral nervous systems. Arch Clin Biomed Res 5 (2021): 484-518.
- Tanaka M, Tanaka T, Teng F, et al. Huaier inhibits cancer progression and induces tissue regeneration by transcriptional regulation of pluripotency of stem cells. J Altern Complement Integr Med 7 (2021): 162-172.
- Tanaka M, Tanaka T, Teng F, et al. Huaier Inhibits Cancer Progression Correlated with the Mutated EGFR and Other Receptor Tyrosine Kinases (c-MET/erbB-2) by Down-Regulation of Multiple Signal Transduction Pathways. Arch Clin Biomed Res 5 (2021): 262-284.
- Tanaka M, Tanaka T, Teng F, et al. Complete remission of the severe advanced stage cancer by miRNA-mediated transcriptional control of Bcl-xL with Huaier therapy compared to the conventional chemotherapy with platinum (II) complex. Arch Clin Biomed Res 5 (2021): 230-261.
- Tanaka M, Tanaka T, Teng F, et al. Anti-cancer effects of Huaier on prostate cancer; miRNA mediated transcription control induced both inhibition of active progression and prevention of relapse. J Altern, Compl Integr Med 7 (2021): 146-154.
- Tanaka M, Tanaka T, Teng F, et al. Huaier induces cancer recovery by rescuing impaired function of transcription control based on the individual genomic potential. Arch Clin Biomed Res 4 (2020): 817-855.
- Tanaka T, Suzuki T, Nakamura J, et al. Huaier regulates cell fate by the rescue of disrupted transcription control in the hippo signaling pathway. Arch Clin Biomed Res 1 (2017): 179-199.
- Tanaka M, Tanaka T. Huaier Natural Herb Therapy for Cancer, Bradeion Institute of Medical Sciences Press (2022).
- Tanaka M, Tanaka T, Teng F, et al. Huaier inhibits cancer progression and induces tissue regeneration by transcriptional regulation of pluripotency of stem cells. J Altern Complement Integr Med 7 (2021): 162-172.
- Tanaka M, Tanaka T, Zhu, X, et al. Hair growth and restoration by transcriptional control of tissue regeneration in cancer recovery process by Huaier. J Cancer science & clinical therapeutics 6 (2022): 288-315.
- Peng Z, Cheng Y, Tan BC, et al. Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat Biotechnol 30 (2012): 253-260.
- Chen Y, Chen Y, Shi C, et al. SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. GigaScience 7 (2018): 1-6.
- Bo Li, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform 12 (2011): 323.
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15 (2014): 1-21.
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 9 (2012): 357-359.
- Enright AJ, John B, Gaul U, et al. MicroRNA targets in Drosophila. Genome Biol 5 (2003): R1.
- Agarwal V, Bell GW, Nam J, et al. Predicting effective microRNA target sites in mammalian mRNAs. Elife 4 (2015): e05005.
- Gendler SJ, Lancaster CA, Taylor-Papadimitriou J, et al. Molecular cloning and expression of human tumor-associated polymorphic epithelial mucin. J Bio Chem 265 (1990): 15286-15293.
- Hollingsworth MA, Swanson BJ. Mucins in cancer: protection and control of the cell surface. Nat Rev Cancer 4 (2004): 45-60.
- Singh PK, Hollingsworth MA. Cell surface-associated mucins in signal transduction. Trends Cell Biol 16 (2006): 467-476.
- Man HY, Wang Q, Lu WY, et al. Activation of PI3-kinase is required for AMPA receptor insertion during LTP of mEPSCs in cultured hippocampal neurons. Neuron 38 (2003): 611-624.
- King D, Yeomanson D, Bryant HE. PI3King the lock: targeting the PI3K/Akt/mTOR pathway as a novel therapeutic strategy in neuroblastoma. J Pediatr Hematol Oncol 4 (2015): 245-245.
- Peltier J, O'Neill A, Schaffer DV. PI3K/Akt and CREB regulate adult neural hippocampal progenitor proliferation and differentiation. Dev Neurobiol 67 (2007): 1348-1361.
- Rafalski VA, Brunet A. Energy metabolism in adult neural stem cell fate. Prog. Neurobiol 93 (2011): 182-203.
- Ojeda L, Gao J, Hooten KG, et al. Critical role of PI3K/Akt/GSK3β in motoneuron specification from human neural stem cells in response to FGF2 and EGF. PLOS ONE 6 (2011): e23414.
- Wyatt LA, Filbin MT, Keirstead HS. PTEN inhibition enhances neurite outgrowth in human embryonic stem cell-derived neuronal progenitor cells. J Comp Neurol 522 (2014): 2741-255.
- Kanehisa M, Araki M, Goto S, Hattori M, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res 36 (2008): 480-484.
- Zhu X. et al. Comparative gene expression profiling and character development instable angina pectoris disease. J Altern Complement Integr Med 7 (2021): 192.
- Ju Son D. The atypical mechanosensitive microRNA-712 derived from pre-ribosomal RNA induces endothelial inflammation and atherosclerosis. Nat Commun 4 (2013): 3000.
- Sloan KE, Warda AS, Sharma S, et al. Tuning the ribosome: The influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Bio 14 (2017): 1138-1152.
- Cao X, Li W, Wang T, et al. Accelerated biological aging in COVID-19 patients. Nat Commun 13 (2022): 2135.
- Chan JC, Hannan KM, Riddell K, et al. AKT promotes rRNA synthesis and cooperates with c-MYC to stimulate ribosome biogenesis in cancer". Sci Signal 4 (2011): ra56.
- Hoppe S, Bierhoff H, Cado I, et al. AMP-activated protein kinase adapts rRNA synthesis to cellular energy supply. Proc Natl Acad Sci USA 106 (2009): 17781-17786.
- Liang XH, Liu Q, Fournier MJ. Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing. RNA 15 (2009): 1716-1728.
- Brandman O, Hegde RS. Ribosome-associated protein quality control. Nat. Struct. Mol. Biol 23 (2016): 7-15.
- King D, Yeomanson D, Bryant HE. PI3King the lock: targeting the PI3K/Akt/mTOR pathway as a novel therapeutic strategy in neuroblastoma. J Pediatr Hematol Oncol 37 (2015): 245-251.
- Rafalski VA, Brunet A. Energy metabolism in adult neural stem cell fate. Prog. Neurobiol 93 (2011): 182-203.
- Man HY, Wang Q, Lu WY, et al. Activation of PI3-kinase is required for AMPA receptor insertion during LTP of mEPSCs in cultured hippocampal neurons. Neuron 38 (2003): 611-624.
- Wyatt LA, Filbin MT, Keirstead HS. PTEN inhibition enhances neurite outgrowth in human embryonic stem cell-derived neuronal progenitor cells. J Comp Neurol 522 (2014): 2741-255.
- Cohen, J. First self-copying mRNA vaccine proves itself in pandemic trial. Science 376 (2022): 6592.
- Ivashchenko A, Pyrkova A, Akimniyazova A, et al. piRNAs are a new COVID-19-fighting tool. Res Square.
Supplementary Files
Supplementary Extended Table 1: SNPs (upper panel) and splice-type variants (lower panel) by the time course. The patient samples: by the time course of Huaier administration; 1. Before Huaier administration; 2. One month after Huaier administration; 3. 3 months after Huaier administration. Normal controls No. 1 and 2: 1. 9 months after Pfizer-BioNTech 1st mRNA vaccination (and before 3rd vaccine shot); 2. 1 month after the 3rd vaccination; 5 months after the 3rd vaccination. The numbers of RNA editing events identified in each sample by splice event types.
Supplementary Extended Table 2: Comparison of DEGs related to the production of iPS cells (KIT, Myc, Oct 3/4, S0X2, Lin28A and NANOG). a. The red box represents up-regulated molecules, and the blue box represents down-regulated molecules. b. The quantitative changes were indicated by log2 transformed fold change calculation by the red bars (up-regulated molecules) and blue bars (down-regulated ones).


 Impact Factor: * 5.3
Impact Factor: * 5.3 Acceptance Rate: 75.63%
Acceptance Rate: 75.63%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 
