Acute Hepatitis C Virus Infections in Spouses: The Utility of a Genetic Analysis of the Hepatitis C Virus Hypervariable Region Sequence for Identifying the Infectious Source
Hiroshi Okano1*, Masaru Murakami2, Hiroki Asakawa1, Kenji Nose1, Satomi Tsuruga1, Tomomasa Tochio1, Hiroaki Kumazawa1, Takashi Sakuno1, Yoshiaki Isono1, Hiroki Tanaka1, Shimpei Matsusaki1, Tomohiro Sase1, Tomonori Saito1, Katsumi Mukai1, Akira Nishimura1, Hiroshi Ohnishi3, Masaharu Takahashi3, Kazumoto Murata3, Hiroaki Okamoto3
1Departments of Gastroenterology, Suzuka General Hospital, Mie, Japan
2Department of Psychiatry, National Hospital Organization Sakakibara Hospital, Tsu, Japan
3Division of Virology, Department of Infection and Immunity, Jichi Medical University School of Medicine, Tochigi, Japan
*Corresponding Author: Dr. Hiroshi Okano, Departments of Gastroenterology, Suzuka General Hospital, 1275-53 Yasuduka-cho, Suzuka, Mie 513-8630, Japan
Received: 16 June 2021; Accepted: 24 June 2021; Published: 21 July 2021
Article Information
Citation: Hiroshi Okano, Masaru Murakami, Hiroki Asakawa, Kenji Nose, Satomi Tsuruga, Tomomasa Tochio, Hiroaki Kumazawa, Takashi Sakuno, Yoshiaki Isono, Hiroki Tanaka, Shimpei Matsusaki, Tomohiro Sase, Tomonori Saito, Katsumi Mukai, Akira Nishimura, Hiroshi Ohnishi, Masaharu Takahashi, Kazumoto Murata, Hiroaki Okamoto. Acute Hepatitis C Virus Infections in Spouses: The Utility of a Genetic Analysis of the Hepatitis C Virus Hypervariable Region Sequence for Identifying the Infectious Source. Archives of Clinical and Medical Case Reports 5 (2021): 537-548.
View / Download Pdf Share at FacebookAbstract
A married couple who developed hepatitis was referred to our hospital. The husband was diagnosed with acute hepatitis C virus (HCV) infection based on HCV RNA positivity and seroconversion to HCV antibody. The wife was also diagnosed with HCV-related hepatitis; however, she could not be confirmed to have acute hepatitis due to the lack of information on her HCV negativity just before this event. The HCV strains recovered from the couple were genotype 2b and shared 100% identities within the 5’-untranslated region-core region sequence (655 nucleotides/nt) and non-structural (NS)5B region sequence (502 nt). The amplified hypervariable region 1 (HVR-1) sequence indicated that all 10 clones from the wife shared 100% identity and were identical to 3 of 10 heterogeneous clones (separable into 4 groups) from the husband. The husband had a history of intravenous drug use. These results suggested that one of four quasispecies 2b HCV strains was transmitted from the husband to the wife, with the husband being the infectious source for acute HCV infection in the wife, most likely via sexual intercourse. A sequence analysis of the HCV genomes and the further comparison of the HVR-1 amino acid sequence variability may be useful for defining the infectious source of HCV, especially in couples or cluster cases.
Keywords
<p style="text-align:justify">Acute hepatitis C; Hepatitis C virus; Genotype; Hypervariable region; Interspousal transmission</p>
Article Details
1. Introduction
Although hepatitis C virus (HCV) infection remains a major global public health burden, with characteristics of chronicity and serious end-stage conditions, including cirrhosis, decompensation, and hepatocellular carcinoma, the occurrence of acute HCV infection is decreasing [1, 2]. The diagnosis of acute-phase HCV infection is difficult because of its subclinical manifestation and/or non-specific illness [3]. Fulminant hepatitis caused by acute HCV infection is rarely observed [4], and symptomatic individuals are likely to clear the virus [3]. However, most cases of acute HCV infection asymptomatically progress to chronic hepatitis. There are almost no new HCV infections due to blood transfusion, intravenous drug use [3] and iatrogenic transmissions [5-8] as the major causes of acute HCV infection. Although sexual transmission is a minor mode of HCV transmission [9-12], aside from in human immunodeficiency virus (HIV)-infected individuals [10], sexual intercourse has been recognized as an HCV infection route [9-17]. To identify the person serving as the source of an infection among implicated patients, the disease onset time is generally compared [6, 7, 18-20]. While nucleotide sequencing and phylogenetic analyses are most accurate for such purposes, amino acid substitutions in the hypervariable region 1 (HVR-1) of the HCV genome are observed in the acute phase of infection [21-23], and the usefulness of genetic diversity of HVR variations for the detection of recent or chronic HCV infections has been reported [24]. We herein report a rare case of interspousal HCV transmission that occurred in spouses who were simultaneously diagnosed with HCV-related hepatitis and whose source was determined to be the husband based on the results of sequence variability of the HCV HVR-1 clones.
2. Case Report
A married couple with hepatitis was referred to our hospital.
The husband was 46 years old, and the wife was 33 years old. The husband had been diagnosed with dilated cardiomyopathy one year earlier at Yokkaichi Municipal Hospital, and his history included psychophysiologic disorder. Approximately two months prior to the onset of hepatitis, the husband had been referred to his primary care physician by Yokkaichi Municipal Hospital to follow-up his cardiomyopathy, and he had continued taking his medications, including anticoagulant therapy. At the initial follow-up visit of his primary care physician, his laboratory data showed a normal hepatic function, and antibody against HCV (HCVAb) was negative. However, elevated hepatic enzymes (alanine aminotransferase [ALT] 315 IU/L; aspartate aminotransferase [AST] 446 IU/L) were noted approximately 2 months after the first follow-up visit of his primary care physician. The wife had undergone a pregnancy workup approximately 15 months before the hepatitis onset and had been HCVAb-negative at this time. Her history included dissociative disorder, for which she had been followed by a psychiatrist since 21 years old. When the husband received regular laboratory workup and was found to have elevated hepatic enzymes (ALT 315 IU/L; AST 446 IU/L) by his primary care physician, she also simultaneously underwent laboratory testing because of complaints of general fatigue, with her laboratory data indicating elevated hepatic enzymes (ALT 379 IU/L; AST 369 IU/L).
Table 1 shows the data of both spouses at the first visit to our hospital. Both patients had elevated hepatic enzyme levels and were positive for HCVAb and HCV RNA. The wife’s laboratory results indicated positive IgM antibodies against herpes simplex virus and cytomegalovirus, but these positive reactions were assumed to be non-specific reactions because she had no fever and no common cold symptoms. We consequently diagnosed the husband with acute HCV infection because of his elevated hepatic enzymes, HCVAb positivity, and HCV RNA positivity, even though a previous test showed HCVAb negativity. We also diagnosed the wife with HCV infection, but we could not confirm an acute infection because we had no evidence concerning her HCV infection status from six months prior to the hepatitis onset. The HCV infection did not spontaneously clear in either patient, and both patients continued to demonstrate serum HCV RNA positivity and elevated hepatic enzymes at three months after the first visit. Therefore, we diagnosed them with chronic HCV infection and treated them with a direct-acting antiviral agent (DAA). These DAA treatments were approved by the ethics committee of Suzuka General Hospital (Ethics Committee Approval Number 259). The patients completed 12 weeks of sofosbuvir/ribavirin (SOF/RBV) therapy without tapering, interruption, or mental status changes. Their serum HCV RNA content became negative after 4 weeks starting DAA therapy, and their hepatic enzyme levels normalized. HCV RNA negativity continued for 24 weeks after the end of treatment, and the patients achieved sustained virologic responses (SVRs). With regard to their HCV infectious source, both patients initially denied intravenous drug use, a blood transfusion history, sharing of razors or toothbrushes, or having sexual intercourse with someone other than each other. However, when the wife was admitted to the National Hospital Organization Sakakibara Hospital for treatment of dissociative disorder after achieving an SVR, the husband confessed to us that he had secretly started injecting himself with drugs several months before the hepatitis onset. This information prompted us to consider that the husband might have been infected with HCV first, with interspousal HCV transmission then occurring from the husband to the wife.
|
|
Reference |
Husband |
Wife |
|
CBC |
|||
|
WBC (/μl) |
3500-9100 |
6900 |
7300 |
|
RBC (x 104/μl) |
376-500 |
544 |
408 |
|
Hemoglobin (g/dl) |
11.3-15.2 |
16.7 |
10.7 |
|
Hematocrit (%) |
33.4-44.9 |
48.0 |
31.5 |
|
Platelets (x 104/μl) |
13.0-36.9 |
22.5 |
24.7 |
|
Coagulation |
|||
|
PT (%) |
70-130 |
40a |
100 |
|
PT-INR |
0.91-1.14 |
1.67a |
1.0 |
|
Chemistry |
|||
|
AST (IU/L) |
10-35 |
621 |
314 |
|
ALT (IU/L) |
10-35 |
412 |
355 |
|
LDH (IU/L) |
110-225 |
494 |
177 |
|
ALP (IU/L) |
229-520 |
321 |
359 |
|
γ-GT (IU/L) |
8-60 |
584 |
55 |
|
T-Bil (mg/dl) |
0.2-1.3 |
1.7 |
0.7 |
|
D-Bil (mg/dl) |
0.1-0.5 |
0.7 |
0.2 |
|
IgG (mg/dl) |
870-1700 |
2024 |
1273 |
|
IgA (mg/dl) |
110-410 |
402 |
203 |
|
IgM (mg/dl) |
35-220 |
60 |
371 |
|
Viral markers |
|||
|
HBsAg (COI) |
<1.0 |
0.1 (-) |
0.2 (-) |
|
IgM-HBcAb (S/CO) |
<1.0 |
0.04 (-) |
0.1 (-) |
|
IgG-HBcAb (COI) |
<1.0 |
21.9 (+) |
0.1 (-) |
|
HBV DNA (logIU/ml) |
<1.0 |
<1.0 (-) |
NT |
|
HCVAb (COI) |
<1.0 |
12.7 (+) |
7.6 (+) |
|
HCV RNA (logIU/ml) |
<1.2 |
7.6 (+) |
5.4 (+) |
|
IgM-HAVAb (S/CO) |
<0.8 |
0.1 (-) |
0.3 (-) |
|
IgA-HEVAb |
(-) |
(-) |
(-) |
|
IgM-HSVAb |
<0.8 |
<0.8 (-) |
7.22 (+) |
|
IgM-CMVAb (S/CO) |
<0.8 |
<0.8 (-) |
3.08 (+) |
|
IgM-EBVAb |
<10 |
<10 (-) |
<10 (-) |
|
EBNA |
<10 |
10 (+) |
80 (+) |
|
HIV-1 RNA |
(-) |
(-) |
(-) |
|
Immunochemistry |
|||
|
ANA |
<40 |
<40 (-) |
40 (+) |
|
AMAM2 |
<7.0 |
<1.5 (-) |
2.1 (-) |
CBC, complete blood count; WBC, white blood cells; RBC, red blood cells; PT, prothrombin time; PT-INR, prothrombin time-international normalized ratio; AST, aspartate aminotransferase; ALT, alanine aminotransferase; LDH, lactate dehydrogenase; ALP, alkaline phosphatase; g-GT, g-glutamyltransferase; T-Bil, total bilirubin; D-Bil, direct bilirubin; Ig, immunoglobulin; HBsAg, hepatitis B surface antigen; COI, cut-off index; Ab, antibody; HBc, hepatitis B core; S/CO, signal/cut-off; HCV, hepatitis C virus; HAV, hepatitis A virus; HEV, hepatitis E virus; HSV, herpes simplex virus; CMV, cytomegalovirus; EBV, Epstein-Barr virus; EBNA, Epstein-Barr virus nuclear antigen; HIV-1, human immunodeficiency virus-1; ANA, antinuclear antibody; AMAM2, anti-mitochondrial M2 antibody; NT , not tested. a under anti-coagulation treatment.
Table 1: Laboratory data obtained at the first visit to our hospital.
To verify the interspousal transmission, we determined the genotype of each HCV strain and compared the genetic sequences according to the previously described method [25]. RNA extracted from the sera of both spouses was subjected to nested reverse transcription (RT)-polymerase chain reaction (PCR), and direct sequencing of the amplicons were performed. Using the HCV-5’-untranslated region-core region (655 nucleotides [nt]) and HCV non-structural (NS)5B region (502 nt), HCV genotype 2b infection was confirmed by a phylogenetic tree analysis with the neighbor-joining method (Figure 1). The nucleotide sequence homology of both 655 nt in the 5’ untranslated region (UTR)-core region and 502 nt in the NS5B region was 100%. When compared with the reported HCV strain with the highest similarity retrievable from DDBJ/EMBL/GenBank databases as of March 2021, the patients’ HCV strains shared only 98.5% identity within the 5’UTR-core region sequence (Figure 1A) and 95.8% within the NS5B region sequence (Figure 1B). A comparison among the HCV sequences from the couple revealed interspousal HCV infection.
Next, to verify the direction of interspousal transmission, RT-PCR of HVR-1 was conducted on the HCV strains isolated from each patient, and the amplified products were molecularly cloned according to the previously described method [26]. Ten clones were obtained from each patient and subjected to an analysis of the HVR-1 variability. At the amino acid level, the 10 clones from the husband were segregated into 4 groups, with marked sequence divergence in HVR-1 (Figure 2). Five clones in 1 group had at most 11 amino acid substitutions (11/25: 44%), compared with 5 clones that belonged to 3 other groups. In contrast to the marked sequence divergence in HVR-1 in the HCV clones obtained from the husband, all 10 clones from the wife showed 100% amino acid sequence identity in HVR-1 among each other (Figure 2). The comparison of the HVR-1 amino acid sequence among clones from each spouse indicated that all 10 clones from the wife were 100% identical to 3 of the 10 clones from the husband, which formed 1 group (Figure 2). These HVR-1 sequence analysis findings suggested interspousal transmission of HCV from the husband to the wife. Based on the HCV infection-associated risk behavior performed by the husband and the results of the genetic analysis of HCV HVR-1, we finally concluded the couple’s acute HCV infection to be due to preceding infection with HCV in the husband through intravenous drug use, followed by interspousal transmission from the husband to his wife (Figure 3).
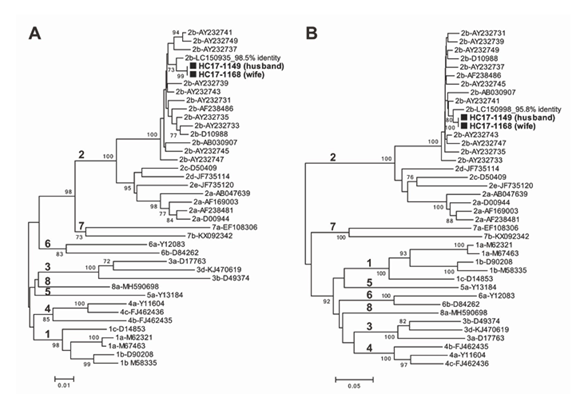
Figure 1: Phylogenetic trees constructed by the neighbor-joining method based on the 655-nt 5’UTR-core sequence (A) and 502-nt NS5B sequence (B) of 40 HCV strains. In addition to the two HCV strains (HC17-1149 from the husband and HC17-1168 from the wife) obtained in the present study, 38 representative HCV strains of genotypes 1a–8a, indicated by genotype and accession number, were included for comparison. The two HCV strains obtained in the present study are highlighted by closed boxes and indicated in bold. The bootstrap values (≥70%) are presented as the percentage of data from 1000 resampling analyses. The scale bars indicate the number of nucleotide substitutions per site. The nucleotide sequence data determined in the present study have been deposited in the DDBJ/EMBL/GenBank databases under accession numbers LC613014-LC613017.

Figure 2: Amino acid sequences of the envelope 1 (E1) and envelope 2 (E2) junctional regions, including hypervariable region 1 (HVR-1), in HCV clones obtained from plasma samples from both patients (husband and wife). The cDNA/PCR clone was propagated in the plasma from both patients at the first visit to our hospital. cDNA/PCR products were subcloned, and 10 clones from each patient were then sequenced. The predicted amino acid sequences of the 10 clones each from the wife and husband are shown. The amino acid positions indicated at the top are in accordance with the prototype genotype 2b HCV strain (D10988). Dots indicate the amino acids identical to the top sequence. The HVR-1 sequences are boxed.
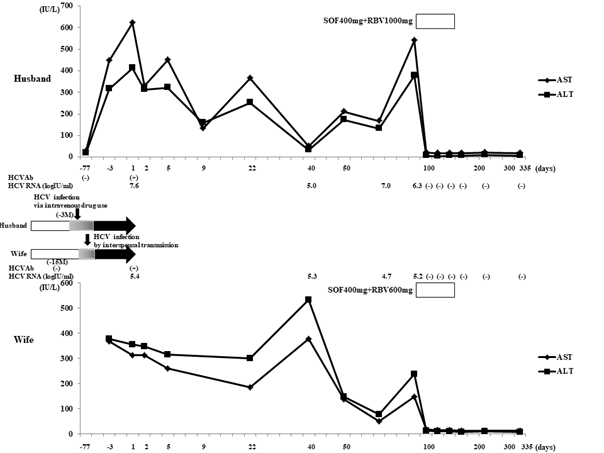
Figure 3: Clinical course and temporal sequences of the proposed transmission route of HCV from the husband to the wife. Day 1: timepoint of the first visit to our hospital with complaints of hepatitis. Open columns indicate HCV RNA negative terms. Closed arrows indicate HCV RNA positive terms. Gray columns indicate the terms presumed to be infected with HCV. The reason we estimated the onset of infection to be three months ago is based on the fact that the husband was negative for HCVAb two months before hepatitis onset.
3. Discussion
Although rapid and effective progress in the treatment of chronic HCV infection have been achieved, HCV infection and ongoing transmission rates remain high within specific populations, especially the major risk group of intravenous drug users [27]. In the present case, the husband was suspected of having contracted HCV through intravenous drug use, and his HCV infection showed the typical pattern of acute HCV infection: HCV seroconversion after two months, serum HCV RNA positivity, and an asymptomatic condition. While sexual transmission is a minor mode of HCV transmission in HIV-negative individuals, the patients were diagnosed with HCV infection via interspousal sexual transmission based on 100% HCV nucleotide sequence homology in the 5’ UTR-core region and NS5B region. The husband had an HCV RNA level of 7.6 log IU/ml on admission, and his high viral load might have been the reason HCV was sexually transmitted to his wife, as a high concentration of circulating HCV is likely to be transmitted to a sexual partner [28, 29].
Because changes in the HCV HVR-1 sequences occur during the course of chronic infection and genetic diversity in HCV correlates with time since infection [30, 31], we analyzed HVR-1 and compared the amino acid sequences among clones from both patients. All 10 clones from the wife were 100% identical to 3 of the 10 clones from the husband. As Lai et al. [17] reported that the comparison of the variability of HCV HVR-1 is useful for confirming interspousal sexual transmission, our analysis comparing each HVR-1 clone supported interspousal HCV transmission in this couple, as did the comparison of the NS5B region sequence. Interestingly, the 10 clones from the husband formed 4 groups with marked sequence divergence in HVR-1 at the amino acid level, although all 10 clones from the wife had 100% HVR-1 amino acid sequence identity with each other. Concerning the estimated HCV mutation rate, a previous study of chimpanzees experimentally infected with HCV revealed that 8 amino acid changes within HVR-1 had occurred within 8.2 years [30]. Furthermore, the amino acid substitution genetic distance within HVR-1 in acute HCV cases was reported to range from 10.8% to 13.8% [22] or 10.2% to 11.1% [21] or be 8.8% (4/45) [23]. In the present case, 11/25 (44%) amino acid substitutions were detected in 1 group of HCV clones from the husband, but all HCV clones from the wife belonged to 1 group and shared 100% amino acid sequence identity with the group consisting of 3 HCV clones without substitutions from the husband. If HCV had been transmitted from the wife to husband, then it might be within two or three months before the hepatitis onset, as he had been confirmed to be HCVAb negative two months before the hepatitis onset. Based on the aforementioned results from previous reports [21-23, 30], it would be difficult to generate 11 amino acid substitutions in HVR-1 within two months. These sequence analysis findings concerning HCV HVR-1 suggested that the transmission route was from the husband to the wife, so interspousal transmission of HCV in the present case was deemed to have likely occurred from the husband to the wife. This finding is further supported by information provided by the psychiatrist, concerning the husband’s intravenous drug use before the hepatitis onset. While we were unable to confirm whether or not the wife used intravenous drugs, we believe that she did not, as the husband had concealed his drug use from his wife. In addition, as the HCV strains from both patients differed from the known HCV strains by ≥1.5% in the 5’UTR-core region sequence and by ≥4.2% in the NS5B region sequence, it is unlikely that the wife was a secret user of intravenous drugs and contracted HCV from a route other than interspousal transmission.
Our previous study of chimpanzees experimentally infected with HCV showed that HVR-1 quasispecies at the acute phase of HCV infection tended to become more prominent as the HCV viral load in the inoculum increased [32]. Notably, HCV is reportedly present at low concentrations in the semen of HCV-positive patients [33], and that the HVR-1 sequence in cases of acute HCV infection via sexual transmission is homogeneous [16]. However, a high level of viral quasispecies was found in individuals who contracted acute-phase HCV infection via blood transfusion, suggesting the occurrence of acute-phase HCV infection with multiple genetic variability in intravenous drug users [34]. These reports support our observation that the HCV HVR-1 sequences in the husband, who was presumed to have contracted HCV infection via intravenous drug use, were heterogeneous, while those in the wife who presumably developed acute HCV infection by interspousal transmission (most likely via sexual intercourse) were homogeneous.
In conclusion, we experienced two cases of acute HCV infection in a married couple. A comparison of the HCV sequences in the NS5B region and 5’UTR-core region revealed that the acute HCV infection had resulted from interspousal transmission. Furthermore, an amino acid sequence analysis of HCV HVR-1 revealed the HCV transmission route to be from husband to wife. Therefore, an analysis and comparison of HVR-1 amino acid sequences may be useful for identifying the infectious source or primary case-patient for HCV outbreaks in a couple or among a group of individuals.
Conflicts of Interest
The authors declare that they have no Conflict of Interest (COI).
References
- Alter MJ. The epidemiology of acute and chronic hepatitis C. Clin Liver Dis 1 (1997): 559-568.
- Yoshizawa H. Hepatocellular carcinoma associated with hepatitis C virus infection in Japan: projection to other countries in the foreseeable future. Oncology 62 (2002): 8-17.
- Westbrook RH, Dusheiko G. Natural history of hepatitis C. J Hepatol 61 (2014): S58-S68.
- Kanzaki H, Takaki A, Yagi T, et al. A case of fulminant liver failure associated with hepatitis C virus. Clin J Gastroenterol 7 (2014): 170-174.
- Griveas I, Germanidis G, Visvardis G, et al. Acute hepatitis C in patients receiving hemodialysis. Ren Fail 29 (2007): 731-736.
- Cody SH, Nainan OV, Garfein RS, et al. Hepatitis C virus transmission from an anesthesiologist to a patient. Arch Intern Med 162 (2002): 345-350.
- Toda T, Mitsui T, Tsukamoto Y, et al. Molecular analysis of transmission of hepatitis C virus in a nurse who acquired acute hepatitis C after caring for a viremic patient with epistaxis. J Med Virol 81 (2009): 1363-1370.
- Ishikawa T, Fukushima Y, Shiobara Y, et al. Outbreak of hepatitis C virus infection in an outpatient clinic. J Gastroenterol Hepatol 20 (2005): 1087-1093.
- Blackard JT, Shata MT, Shire NJ, et al. Acute hepatitis C virus infection: a chronic problem. Hepatology 47 (2008): 321-331.
- Tohme RA, Holmberg SD. Is sexual contact a major mode of hepatitis C virus transmission? Hepatology 52 (2010): 1497-1505.
- Terrault NA, Dodge JL, Murphy EL, et al. Sexual transmission of hepatitis C virus among monogamous heterosexual couples: the HCV partners study. Hepatology 57 (2013): 881-889.
- Kao JH, Liu CJ, Chen PJ, et al. Low incidence of hepatitis C virus transmission between spouses: a prospective study. J Gastroenterol Hepatol 15 (2000): 391-395.
- Nishimura N, Isoda N, Higashizawa T, et al. A case of acute hepatitis C caused by interspousal transmission after 30 years of marriage. Clin J Gastroenterol 3 (2010): 50-56.
- Irving WL, Salmon D, Boucher C, et al. Acute hepatitis C virus infection. Euro Surveill 13 (2008): 18879.
- Nakamura I, Tanaka Y, Ochiai K, et al. Clarification of interspousal hepatitis C virus infection in acute hepatitis C patients by molecular evolutionary analyses: Consideration on sexual and non-sexual transmission between spouses. Hepatol Res 41 (2011): 838-845.
- Nakayama H, Sugai Y, Ikeya S, et al. Molecular investigation of interspousal transmission of hepatitis C virus in two Japanese patients who acquired acute hepatitis C after 40 or 42 years of marriage. J Med Virol 75 (2005): 258-266.
- Lai KW, Young KC, Cheng PN, et al. Interspousal transmission of hepatitis C virus: application of comparing the variability of HVR1 nucleotide region. Hepatogastroenterology 51 (2004): 791-795.
- Schmid D, Fretz R, Kuo HW, et al. An outbreak of multidrug-resistant tuberculosis among refugees in Austria, 2005-2006. Int J Tuberc Lung Dis 12 (2008): 1190-1195.
- Furuse Y, Sando E, Tsuchiya N, et al. Clusters of Coronavirus Disease in Communities, Japan, January-April 2020. Emerg Infect Dis 26 (2020): 2176-2179.
- Brennan J, Mullins H, Tobey K, et al. Nosocomial hepatitis A outbreak among healthcare workers and patients in a community hospital during an ongoing statewide outbreak. Infect Control Hosp Epidemiol 42 (2021): 139-141.
- Higashi K, Tsukiyama-Kohara K, Tanaka T, et al. Characterization of hypervariable region in hepatitis C virus envelope protein during acute and chronic infection. Arch Virol 150 (2005): 883-898.
- Manzin A, Solforosi L, Petrelli E, et al. Evolution of hypervariable region 1 of hepatitis C virus in primary infection. J Virol 72 (1998): 6271-6276.
- Enomoto N, Sakamoto N, Kurosaki M, et al. The hypervariable region of the HCV genome changes sequentially during the progression of acute HCV infection to chronic hepatitis. J Hepatol 17 (1993): 415-416.
- Lara J, Teka M, Khudyakov Y. Identification of recent cases of hepatitis C virus infection using physical-chemical properties of hypervariable region 1 and a radial basis function neural network classifier. BMC Genomics 18 (2017): 880.
- Aikawa T, Tsuda F, Ueno C, et al. Comparison of test results of serogrouping and core region PCR-based genotyping in patients with chronic hepatitis C virus infection: Analysis of inderminate or discrepant cases and identification of a 2b/1b recombinant HCV. Kanzo 57 (2016): 447-456.
- Okada S, Akahane Y, Suzuki H, et al. The degree of variability in the amino terminal region of the E2/NS1 protein of hepatitis C virus correlates with responsiveness to interferon therapy in viremic patients. Hepatology 16 (1992): 619-624.
- Hullegie SJ, Arends JE, Rijnders BJA, et al. Current knowledge and future perspectives on acute hepatitis C infection. Clin Microbiol Infect 21 (2015): 797.e9-797.e17.
- Chayama K, Kobayashi M, Tsubota A, et al. Molecular analysis of intraspousal transmission of hepatitis C virus. J Hepatol 22 (1995): 431-439.
- Yazdanpanah Y, De Carli G, Migueres B, et al. Risk factors for hepatitis C virus transmission to health care workers after occupational exposure: a European case-control study. Clin Infect Dis 41 (2005): 1423-1430.
- Okamoto H, Kojima M, Okada S, et al. Genetic drift of hepatitis C virus during an 8.2-year infection in a chimpanzee: variability and stability. Virology 190 (1992): 894-899.
- Carlisle LA, Turk T, Metzner KJ, et al. HCV genetic diversity can be used to infer infection recency and time since infection. Viruses 12 (2020): 1241.
- Kojima M, Osuga T, Tsuda F, et al. Influence of antibodies to the hypervariable region of E2/NS1 glycoprotein on the selective replication of hepatitis C virus in chimpanzees. Virology 204 (1994): 665-672.
- Cassuto NG, Sifer C, Feldmann G, et al. A modified RT-PCR technique to screen for viral RNA in the semen of hepatitis C virus-positive men. Hum Reprod 17 (2002): 3153-3156.
- Herring BL, Tsui R, Peddada L, et al. Wide range of quasispecies diversity during primary hepatitis C virus infection. J Virol 79 (2005): 4340-4346.


 Impact Factor: * 5.3
Impact Factor: * 5.3 Acceptance Rate: 75.63%
Acceptance Rate: 75.63%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 
