Familial Hemiplegic Migraine: A New Gene in an Italian Family
Pietro Palumbo1#, Orazio Palumbo1#*, Francesca Felicia Operto2, Stefano Castellana3, Ester Di Muro1,4, Maria Pia Leone1, Tommaso Biagini3, Grazia Pastorino2, Tommaso Mazza3, Massimo Carella1*, Giangennaro Coppola2
#Both the authors contributed equally
1Division of Medical Genetics, Fondazione IRCCS Casa Sollievo della Sofferenza, Viale Padre Pio, 71013 San Giovanni Rotondo (FG), Italy
2Child and Adolescent Neuropsychiatry, Department of Medicine, Surgery and Odontoiatry, University of Salerno, Largo d’Ippocrate 1, 84100 Salerno, Italy
3Bioinformatics Unit, Fondazione IRCCS Casa Sollievo della Sofferenza, Viale Padre Pio, 71013 San Giovanni Rotondo (FG), Italy
4Department of Cellular Biotechnologies and Hematology, University of Roma “La Sapienza”, Roma, Italy
*Corresponding Authors: Dr. Orazio Palumbo, Division of Medical Genetics, Fondazione IRCCS Casa Sollievo della Sofferenza, Viale Padre Pio, 71013 San Giovanni Rotondo (FG), Italy
Dr. Massimo Carella, Division of Medical Genetics, Fondazione IRCCS Casa Sollievo della Sofferenza, Viale Padre Pio, 71013 San Giovanni Rotondo (FG), Italy
Received: 06 August 2019; Accepted: 30 August 2019; Published: 06 September 2019
Article Information
Citation: Palumbo P, Palumbo O, Operto FF, Castellana S, Muro ED, Leone MP, Biagini T, Pastorino G, Mazza T, Carella M, Coppola G. Familial Hemiplegic Migraine: A New Gene in an Italian Family. Archives of Clinical and Medical Case Reports 3 (2019): 534-543.
View / Download Pdf Share at FacebookAbstract
Hemiplegic migraine (HM) is a rare form of migraine characterized by severe attacks of unilateral and pulsating headache (associated with photophobia, phonophobia or nausea). To date, mutations in three different genes, ATP1A2, CACNA1A and SCN1A, are known to cause familial hemiplegic migraine (FHM) or sporadic hemiplegic migraine (SHM) phenotype, while mutations in ATP1A3 were recently associated to alternating hemiplegia of childhood (AHC). Here we report an Italian family who presented with familial hemiplegic migraine, carrier of an heterozygous ATP1A4 mutations c.1798 C>T, predicted to cause the p. (Pro600Ser) amino acid substitution, which segregated with the disease phenotype in four members. The variant, identified by using whole exome sequencing combined with Sanger sequencing, was predicted to be disease-causing by prediction software and to affect protein stability by thermodynamic study. To our knowledge, this is the first report of a mutation of ATP1A4 gene, associated to FHM, thus useful to further expand our understanding of the molecular etiology of HM. Furthermore, our evidence may suggest to consider ATP1A4 in addition to ATP1A2, ATP1A3, CACNA1A and SCN1A, in genetic screening of patients affected by HM.
Keywords
<p>ATP1A4; Whole exome sequencing; Hemiplegic migraine</p>
Article Details
Abbreviations:
HM: hemiplegic migraine; FHM: familiar hemiplegic migraine; SHM: sporadic hemiplegic migraine; AHC: alternating hemiplegia of childhood; WES: whole exome sequencing; EKG: ElectrocardiographySVT: supraventricular tachycardia; VT: ventricular tachycardia; MRI: Magnetic Resonance Imaging; EEG: electroencephalography; ACMG: American college of medical genetics
1. Introduction
Hemiplegic migraine (HM) is a rare form of migraine characterized by severe attacks of unilateral and pulsating headaches (associated with photophobia, phonophobia or nausea). Headaches are often preceded by neurological symptoms of aura (visual, sensory, speech and hemiparesis disorders) which can be very prolonged and severe in some patients [1]. HM represents a clinically heterogeneous disorder, which may present additional manifestation such as permanent ataxia, infantile convulsions, epileptic seizures, cerebral edema, and coma after minor head trauma and intellectual disability. In addition, HM may arise as: i) familial hemiplegic migraine (FHM) when at least one first- or second-degree patients’ relative is affected; ii) sporadic hemiplegic migraine (SHM), with the same clinical manifestation of FHM, but no relative presents the disease [2]. Another clinical entity related to HM is the alternating hemiplegia of childhood (AHC), a complex syndrome characterized by episodes of hemiplegia alternating in laterally, paroxysmal disorders, permanent neurological dysfunction, onset of the neurological symptoms before 18 months of age [3].
It is known that the primary cause of familial hemiplegic migraine (and common migraine forms) resides in the brain, but the mechanisms of primary brain dysfunction at the cell and neuronal circuitry are not clearly understood [4]. From a molecular point of view, HM has been described as a monogenic, genetically heterogeneous channelopathy disorder, with four causative genes to date known, CACNA1A (FHM) [5], ATP1A2 (FHM) [6], SCN1A (FHM) [7] and ATP1A3 (AHC) [8].
Despite their role in individual families with several affected family members, mutations in these genes do not seem clarify the majority of cases of HM in the general population, suggesting additional causal or contributing variants/genes for HM. A few other genes have been suggested, but their causality has not been confirmed [9]. In this paper we describe a missense variant in ATP1A4 identified by Whole Exome Sequencing (WES) combined to Sanger sequencing in four members of an Italian non-consanguineous family, affected by FHM, useful to expand our knowledge about the pathogenesis of this condition.
2. Case Presentation
The pedigree of the family described in this report is shown in Figure 1.
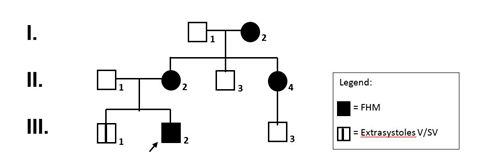
Figure 1: Pedigree of the family. Filled symbols represent affected individuals.
2.1 I.2: female, 76 years of age
Soon after the first pregnancy at the age of 23 years, onset of bilateral headache with an average duration of about 1-2 hours and variable intensity, not always requiring drug therapy. Migraine attacks were preceded by a visual aura lasting about 20 minutes, mainly triggered by sleep deprivation, poor food intake and psychophysical stress. In the last months, headache became frequent (about twice a day, every other day) and severe, associated with visual aura and followed by short lasting limb weakness. Headache was promptly stopped by ibuprofen 600 mg as single dose. Last 24-h holter EKG disclosed 421 supraventricular extrasystolic beats (SVEBs) with twelve couples and 9 short runs.
2.2 II.2: female, 50 years of age
Since the age of 6 years, migraine attacks recurring mainly on the right side with ipsilateral hemianopsia or scintillating scotomas, and subsequent paresthesia at the right upper limb, then at the right side of the tongue and face, and at the ipsilateral lower limb. Each episode lasted at least 2 hours, followed by vomiting, photo- and phonophobia. Confusion and weakness persisted until the next day. An early 600 mg single dose of ibuprofen usually controlled symptoms. In adolescent age about 3-4 extremely severe episodes per year were reported. In the last 20 years (after the first pregnancy), about 1 episode per year until the age of 49, when episodes increased in frequency and intensity. A recent 24-h holter EKG disclosed 7 single SVEBs. Past clinical history: reported diagnosis of psoriatic arthritis, fibromyalgia and severe urolithiasis.
2.3 II.4: female, 46 years of age
Headache onset was at about 12 years of age, with flashing lights in the right eye lasting about half an hour followed by paresthesias and hyposthenia at the right upper limb and ipsilateral labial angle. The migraineous attacks were described as intense, pulsating and disabling. Ketoprofen single dose could stop the progression of symptoms if taken at the onset of the headache. Initially, the frequency of the episodes was 3-4 per year, falling to 1/year until 41 years of age. More recently, sporadic short lasting episodes of hemiplegic migraine tend to recur. A recent 24-h holter EKG recording detected 39 monomorphic isolated ventricular exstrasystolic beats (VEBs), and 18 single SVEBs. Past medical history showed a Sjögren Syndrome and mild urolithiasis.
2.4 III.2: male, proband, 14 years of age
The child was born at term, after an uneventful pregnancy and normal delivery. Newborn body-weight was 3100 g. Psychomotor development and school performances were in the normal range. At the age of 9 years and 4 months, the child manifested upon awakening his first episode of headache, consisting of frontal pulsating pain lasting one hour and a half, associated with central flashing scotomas, paresthesias at the right side, with dysarthria lasting about 30 minutes and subsequent hyposthenia at the same body side. After resolution of these symptoms, the patient made a full recovery. No nausea or vomiting were reported. On the same day he performed an EEG recording and a brain CT scan, both of which were normal. About 20 episodes with overlapping disabling symptoms recurred in the following two months, preventing him from going to school.
Considering the clinical and family history, a Familial Hemiplegic Migraine (FHM) was then suspected, for which a high resolution SNP-Array analysis performed in the patient and his parents showed no pathological deletions or duplications. In addition, Sanger sequencing of the main genes, so far associated with this condition, including CACNA1A, ATP1A2, SCN1A, PRRT2, SLC1A4, SLC4A4, did not disclose any significant mutation. In the last 12 months (age 14 years) headache attacks decreased to 1-2 per month, lasting 2-3 hours each. They were associated with hemiparesis and right amaurosis and responded to ibuprofen therapy. A holter EKG recording performed at the age of 14 years showed no VY, 1 SVT, and no other significant alterations.
3. Material and Methods
3.1 Whole exome sequencing (WES)
Peripheral blood samples were taken from the patient and their parents, and genomic DNA was isolated by using Bio Robot EZ1 (Quiagen, Solna, Sweden). The quality of DNA was tested on 1% electrophorese agarose gel, and the concentrations was quantified with Nanodrop 2000 C spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The molecular testing carried out in this report is based on routine clinical care and local Ethics committee approval was not requested. Proband’s DNA was analyzed by whole exome sequencing (WES) by using SureSelect Human Clinical Research Exome (Agilent Technologies) following manufacturer instructions. This is a combined shearing free transposase-based library prep and target enrichment solution, which enables comprehensive coverage of the entire exome. This system enables a specific mapping of reads to targets for deep coverage of targets protein coding regions from RefSeq, GENCODE, CCDS, and UCSC known Genes, with excellent overall exonic coverage and increased coverage of HGMD, OMIM, ClinVar, and ACMG targets. Sequencing was performed on a NextSeq 500 system (Illumina Inc.) by using the High Output flow cells (300 cycles), with a minimum expected coverage depth of 70x. The average coverage obtained was 107x. Putative pathogenic variant was confirmed by Sanger sequencing in the proband’s and parents’ DNA. PCR products were sequenced by using BigDye Terminator v1.1 sequencing kit (Applied Biosystems) and ABI Prism 3100 Genetic Analyzer (Thermo Fisher Scientific).
3.2 Bioinformatics analysis
The generated paired-end reads were initially checked for their quality with the FastQC tool [Andrews S. (2010). FastQC: a quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc] and then mapped to the hg19 reference genome sequence by means of Bowtie. Depth of coverage statistics for the target regions were calculated by TEQC ver. 3.47. Variants were called by means of the HaplotypeCaller tool of GATK ver. 3.58 [10] and were annotated with ANNOVAR [11], using RefSeq gene and transcript annotations (updated to Dec 2016). Variants were found in dbSNP ver. 150 [12], ExAC ver. 0.3 [13], and Exome Variant Server (http://evs.gs.washington.edu/EVS, accessed at December 2016) [14], HRC [15], Kaviar [16] and ClinVar [17]. Missense variants were further annotated by querying the dbNSFP ver. 3.2 resources and retrieving pre-computed pathogenicity predictions and evolutionary conservation measures [18].
The stability of the ATP1A4 protein upon mutation was investigated thermodynamically through the FoldX16 algorithm (see Results). This study was conducted on the wild-type protein model obtained from the Protein Data Bank [19]. It was mutated in-silico through UCSC Chimera [20], yielding a second protein model. Both were minimized, namely all the side chains were slightly moved in order to reduce the Van der Waals’ clashes, before being analyzed by FoldX. The standalone version of FoldX is downloadable from http://foldx.crg.es. It was run with standard parameters and used to compute the total energy values of the wild-type and mutated models of ATP1A4. These values were used to predict the overall protein stability, with and without mutations.
4. Results
WES allow us to detect an heterozygous missense variant in exon 12 of ATP1A4 (NM_144699) c.1798 C>T, predicted to cause the p.(Pro600Ser) amino acid substitution. This result was confirmed by Sanger sequencing of proband’s and parents’ DNA. The variant, inherited from the affected mother, is present in all affected family members, absent in healthy family members (Figure 2) and it affects a highly conserved residue (GERP++_RS score: 4.2; phyloP100way_vertebrate score: 7.9; phyloP100way_mammalian score: 0.935; phastCons100way_vertebrate score: 1,000; SiPhy_29way_logOdds: 14.110).
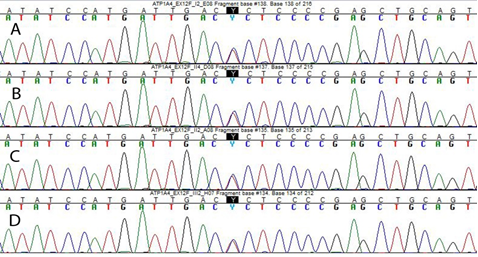
Figure 2: Sanger sequencing showing the c. 1798 C>T nucleotide change in ATP1A4. (A) Electropherogram of the patient I.2, (B) II.4, (C) II.2 and (D) III.2.
Furthermore, the variant is located in the ATP1A4 topological domain and it is predicted to be harmful by several pathogenicity predictor software, including SIFT, PolyPhen2, LRT, CADD, DANN, FatHmm. The variant has been reported in dbSNP (rs142338502), and is quite rare (MAF of 0.001 reported in genomAD and 1000Genomes, 0.002 in ExAC). The free energy calculations for this variant were ΔGmut=44.88 kcal/mol and ΔGwt=42.79 kcal/mol, from which ΔΔG = ΔGmut - ΔGwt = +2.09 kcal/mol. The difference in free energy resulted to be positive and in the range to classify the p.(Pro600Ser) variant as highly destabilizing. The localization of the variant is depicted in Figure 3.
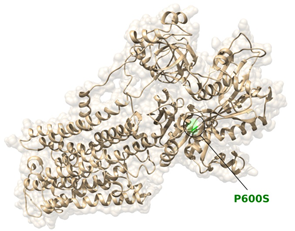
Figure 3: Location of the Pro600Ser substitution in the ATP1A4 protein / Ribbon diagram of ATP1A4, with Pro600Ser colored in green.
5. Discussion
In recent years, increasing evidences showed that HM tends to run in families, suggesting that genetic determinants play a significant role in the disease. It has been shown that at least 50% of migraineurs have a parent affected by a similar condition and a familial liability has been confirmed in several studies [21]. Although familial does not necessarily mean genetic, epidemiological evidence seems to indicate a close gene–environment interaction. To date, four genes, two encoding for ion channels and two encoding an ATP excharger, have been found to be responsible for several forms of HM [22, 23]. CACNA1A (OMIM # 601011) was the first gene associated to FHM, and to date more than 70 causative variants have been identified. CACNA1A encodes the α1 subunit of neuronal CaV2.1 (P/ Q-type) voltage-gated calcium channels that are widely expressed throughout the central nervous system [24]. Variants in this channel causes excessive release of the neurotransmitter glutamate, thus altering physiologic conditions [25]. The second gene encoding for a ion channel is SCN1A (OMIM # 182389), which encodes the α1 subunit of neuronal NaV1.1 voltage-gated sodium channels and is a well-known epilepsy gene, with more than 100 truncating and missense variants associated with epilepsy [26]. The first gene encoding an ATP exchanger and associated to HM is ATP1A2 (OMIM # 182340), which encodes the α2 subunit of sodium-potassium pump. To date more than 40 variants involving ATP1A2 are known to cause FHM both without other clinical manifestation (pure FHM) [6] or in addition to other symptoms such as AHC [27], epilepsy [28], intellectual disability [29]. ATP1A2 have also been associated with non-hemiplegic migraine and common migraine [30], providing a strong evidence of the wide genetic and clinical heterogeneity of these conditions. The second gene encoding an ATP excharger and associated, as ATP1A2, to different neurological conditions such as AHC, rapid-onset Dystonia Parkinsonism (RDP) and CAPOS syndrome (cerebellar ataxia, areflexia, pes cavus, optic atrophy, and sensorineural hearing loss), is ATP1A3 (OMIM #182350) which encodes the α3 subunit of sodium-potassium pump [12]. We describe the first Italian family to date reported in medical literature, with four members affected by FHM in which a novel putative pathogenic variant in ATP1A4 gene has been identified and segregates in all affected members. To the best of our knowledge, this is the first time the ATP1A4 gene has been associated to this clinical condition, and for this reason useful to expand our knowledge regarding the molecular mechanisms underlying HM.
The protein encoded by ATP1A4 gene belongs to the family of P-type cation transport ATPases, and to the subfamily of Na+/K+ -ATPases. Na+/K+ -ATPase is an integral membrane protein responsible for establishing and maintaining the electrochemical gradients of Na and K ions across the plasma membrane, by the active transport of three sodium ions for two potassium ions across cell membranes [31]. These gradients are essential for osmoregulation, for sodium-coupled transport of a variety of organic and inorganic molecules, and for the electrical excitability of nerve and muscle. This enzyme is composed of two subunits, a large catalytic subunit (alpha) and a smaller glycoprotein subunit (beta). The catalytic subunit (alpha) of Na+/K+ -ATPase is encoded by multiple genes, and ATP1A4gene encodes an α4 subunit, highly expressed in testis, but also in the brain and skeletal muscle [32], showing the properties to associate different beta subunits in different tissues [33]. Although, to date, ATP1A4 was never associated to neurological condition, it encodes for one of the component of the catalytic subunit of the Na+/K+ -ATPase, as ATP1A2 and ATP1A3 genes, already known to be causative of FHM, AHC and other neurologic conditions. Thus, due to these functional evidences, the contribution of ATP1A4 alteration in the onset of HM could be likely. In the family described here, composed by eight people among three generations, all affected members (four) carries the variation in ATP1A4. Even if the detected variant presents a MAF higher than expected for a monogenic disorder, this evidence is not uncommon for conditions which did not affect reproductive fitness of carriers people, such as for example in hypertrophic cardiomyopathy.
From a clinical point of view, they presented paresthesias at the right side and an increasing frequency of HM during the years. The age of onset is similar for II2 (6 years old), II4 (12 years old), III2 (9 years old), such as the duration of 1.5-2 hours, and for all patients headache decrease with Ibuprofen/Ketoprofen assumption. Other clinical manifestations are quite different, such as flashing lights in the right eye (II4) and central flashing scotomas (III2), but this is a frequent evidence in pathologies with high clinical heterogeneity. Understanding the primary brain dysfunction involved in hemiplegic migraine is essential in order to develop more specific and effective drugs for the control of clinical manifestation, and to identify new drug targets, so increasing knowledge of genetic contribution to these diseases is crucial.
6. Conclusions
In conclusion, we described for the first time a putative pathogenic variant of ATP1A4, identified in a family affected of FHM. We suggest, in order to increase the chance to identify the molecular cause of the disease, to screen the sequence of ATP1A4 in addition to CACNA1A, SCN1A, ATP1A2 and ATP1A3in patients affected by HM, or to sequence ATP1A4 in all patients negative to previous genetic screening of other candidate genes.
Acknowledgments
We thank the family for their cooperation.
Conflict of InterestThe authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Author Contributions
PP, OP, MC and GC conceived the study, designed experiments and wrote the manuscript. GC, FFO and GP collected the patients’ clinical information. SC, TB and TM performed the analysis of TRS data and modeling the impact of the reported variant on the three-dimensional structure of the protein. EDM and MPL performed the molecular analysis. PP, OP, EDM and MPL interpreted the results. All authors reviewed and approved the final manuscript.
Funding
This study was supported by a grant of the Italian Ministry of Health (Ricerca Corrente 2019) to Massimo Carella.
Consent for Publication
This study was carried out following the recommendations of the Declaration of Helsinki and the Italian law for biomedical experimentation with written informed consent from all subjects.
References
- Headache Classification Committee of the International Headache Society (IHS). The international classification of headache disorders, 3rd edition (beta version). Cephalalgia 38 (2018) 1-211.
- Russell MB, Ducros A. Sporadic and familial hemiplegic migraine: pathophysiological mechanisms, clinical characteristics, diagnosis, and management. Lancet Neurol 10 (2011): 457-470.
- Panagiotakaki E, De Grandis E, Stagnaro M, et al. Clinical profile of patients with ATP1A3 mutations in Alternating Hemiplegia of Childhood-a study of 155 patients. Orphanet J Rare Dis. 10 (2015): 123.
- Zarcone D, Corbetta S. Shared mechanisms of epilepsy, migraine and affective disorders. Neurol Sci 38 (2017): 73-76.
- Ophoff RA, Terwindt GM, Vergouwe MN, et al. Familial hemiplegic migraine and episodic ataxia type-2 are caused by mutations in the Ca2+ channel gene CACNL1A4. Cell 87 (1996): 543-552.
- De Fusco M, Marconi R, Silvestri L, et al. Haploinsufficiency of ATP1A2 encoding the Na+/K+ pump alpha2 subunit associated with familial hemiplegic migraine type 2. Nat Genet 33 (2003): 192-196.
- Dichgans M, Freilinger T, Eckstein G, et al. Mutation in the neuronal voltage-gated sodium channel SCN1A in familial hemiplegic migraine. Lancet 366 (2005): 371-377.
- Carecchio M, Zorzi G, Ragona F, et al. ATP1A3-related disorders: An update. Eur J Paediatr Neurol 22 (2017): 257-263.
- Ferrari MD, Klever RR, Terwindt GM, et al. Migraine pathophysiology: lessons from mouse models and human genetics. Lancet Neurol 14 (2015): 65-80.
- McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20 (2010): 1297-1303.
- Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from next-generation sequencing data. Nucleic Acids Res 38 (2010): e164.
- Sherry ST, Ward MH, Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 29 (2001): 308-311.
- Exome Aggregation Consortium. Analysis of protein-coding genetic variation in 60,706 humans. BioRxiv (2015). doi: http://dx.doi.org/10.1101/030338
- Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP), Seattle, WA (URL: http://evs.gs.washington.edu/EVS/)
- McCarthy S, Das S, Kretzschmar W, et al. Haplotype Reference Consortium. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet 48 (2016): 1279-1283.
- Glusman G, Caballero J, Mauldin DE, et al. KAVIAR: an accessible system for testing SNV novelty. Bioinformatics 27 (2011): 3216-3217.
- Landrum MJ, Lee JM, Riley GR, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res 42 (2014): D980-D985.
- Liu X, Wu C, Li C, et al. dbNSFP v3.0: A One-Stop Database of Functional Predictions and Annotations for Human Non-synonymous and Splice Site SNVs. Hum Mutat 37 (2016): 235-241.
- Berman HM, Battistuz T, Bhat TN, et al. The Protein Data Bank. Acta Crystallografica D58 (2002): 899-907.
- Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera, A Visualization System for Exploratory Research and Analysis. J Comput Chem 25 (2004): 1605-1612.
- Schu¨rks M. Genetics of migraine in the age of genome wide association studies. J Headache Pain 13 (2012): 1-9.
- Hiekkala ME, Vuola P, Artto V, et al. The contribution of CACNA1A, ATP1A2 and SCN1A mutations in hemiplegic migraine: A clinical and genetic study in Finnish migraine families. Cephalalgia 38 (2018): 1849-1863.
- Carecchio M, Zorzi G, Ragona F, et al. ATP1A3-related disorders: An update. Eur J Paediatr Neurol 22 (2018): 257-263.
- Westenbroek RE, Sakurai T, Elliott EM, et al. Immunochemical identification and subcellular distribution of the alpha 1A subunits of brain calcium channels. J Neurosci 15 (1995): 6403-6418.
- van den Maagdenberg AM, Pietrobon D, Pizzorusso T, et al. A Cacna1a knockin migraine mouse model with increased susceptibility to cortical spreading depression. Neuron 41 (2004): 701-710.
- Vahedi K, Depienne C, Le Fort D, et al. Elicited repetitive daily blindness: a new phenotype associated with hemiplegic migraine and SCN1A mutations. Neurology 72 (2009): 1178-1183.
- Swoboda KJ, Kanavakis E, Xaidara A, et al. Alternating hemiplegia of childhood or familial hemiplegic migraine? A novel ATP1A2 mutation. Ann Neurol 55 (2004): 884-887.
- Jurkat-Rott K, Freilinger T, Dreier JP, et al. Variability of familial hemiplegic migraine with novel A1A2 Na(+)/K(+)-ATPase variants. Neurology 62 (2004): 1857-1861.
- Vanmolkot KR, Stroink H, Koenderink JB, et al. Severe episodic neurological deficits and permanent mental retardation in a child with a novel FHM2 ATP1A2 mutation. Ann Neurol 59 (2006): 310-314.
- Todt U, Dichgans M, Jurkat-Rott K, et al. Rare missense variants in ATP1A2 in families with clustering of common forms of migraine. Hum Mutat 26 (2005): 315-321.
- Woo AL, James PF, Lingrel JB. Characterization of the fourth alpha isoform of the Na,K-ATPase. J Membr Biol 169 (1999): 39-44.
- Lingrel J, Moseley A, Dostanic I, et al. Functional roles of the alpha isoforms of the Na,K-ATPase. Ann N Y Acad Sci 986 (2003): 354-359.
- Sanchez G, Nguyen AN, Timmerberg B, et al. The Na,K-ATPase alpha4 isoform from humans has distinct enzymatic properties and is important for sperm motility. Mol Hum Reprod 12 (2006): 565-576.


 Impact Factor: * 5.3
Impact Factor: * 5.3 Acceptance Rate: 75.63%
Acceptance Rate: 75.63%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 
